Bacteriophages, viruses that infect bacteria, have emerged as a legitimate alternative antibacterial agent with a wide scope of applications which continue to be discovered and refined. However, the potential of some bacteriophages to aid in the acquisition, maintenance, and dissemination of negatively associated bacterial genes, including resistance and virulence genes, through transduction is of concern and requires deeper understanding in order to be properly addressed. In particular, their ability to interact with mobile genetic elements such as plasmids, genomic islands, and integrative conjugative elements (ICEs) enables bacteriophages to contribute greatly to bacterial evolution. Nonetheless, bacteriophages have the potential to be used as therapeutic and biocontrol agents within medical, agricultural, and food processing settings, against bacteria in both planktonic and biofilm environments. Additionally, bacteriophages have been deployed in developing rapid, sensitive, and specific biosensors for various bacterial targets. Intriguingly, their bioengineering capabilities show great promise in improving their adaptability and effectiveness as biocontrol and detection tools.
- bacteriophage
- phage therapy
- antibiotic resistance
1. Introduction
The use of antibiotics to treat a wide range of infections, saving millions of lives and revolutionizing the field of medicine since their discovery, has enabled them to be one of the most impactful scientific discoveries in modern history. However, the resulting consequences of their extensive use has signified a new era for medicine. The rapid rise of antibacterial resistance in bacteria across the globe, coupled with declines in the development of novel antibacterial agents, has resulted in the need for new approaches in combating bacterial infections [1]. Bacteriophages (phages), viruses that infect bacteria, have emerged as a viable alternative to the declining utility of traditional antimicrobials to mitigate the risk of pathogenic bacteria [2]. Their ubiquity in nature, versatility, and innate resiliency has elevated their status from mere research organisms to potentially viable and necessary tools in the fight against rising antimicrobial resistance [3].
While their potential utility and benefits continued to be examined and documented, one of the key hurdles facing the progression and ultimate acceptance of bacteriophages for therapeutic and biocontrol purposes is their ability to contribute to the horizontal transfer of genes, which can have negative consequences. The transfer of resistance, virulence, and other negatively associated genes via bacteriophages is a major area in which further examination and understanding is needed, so that proper steps may be taken to minimize its potentially harmful impacts.
Conversely, bacteriophages provide an incredible sense of versatility for potential application. The use of whole phage particles and phage components continues to be studied, as well as their use across a variety of industries and purposes. The application of bacteriophages within medical, agricultural, and food processing settings provides some of the most promising opportunities for their regulatory approval and commercialization [4][5]. Their ability and potential efficacy to be used against bacteria in both planktonic and biofilm settings provide another reason for their continued examination [6]. Phages continue to be studied as not only bacterial killers, but also as potential active bacterial detectors, and while understanding surrounding them remains relatively limited, computational and bioengineering advancements have the potential to further bolster the utility of the bacterial viruses.
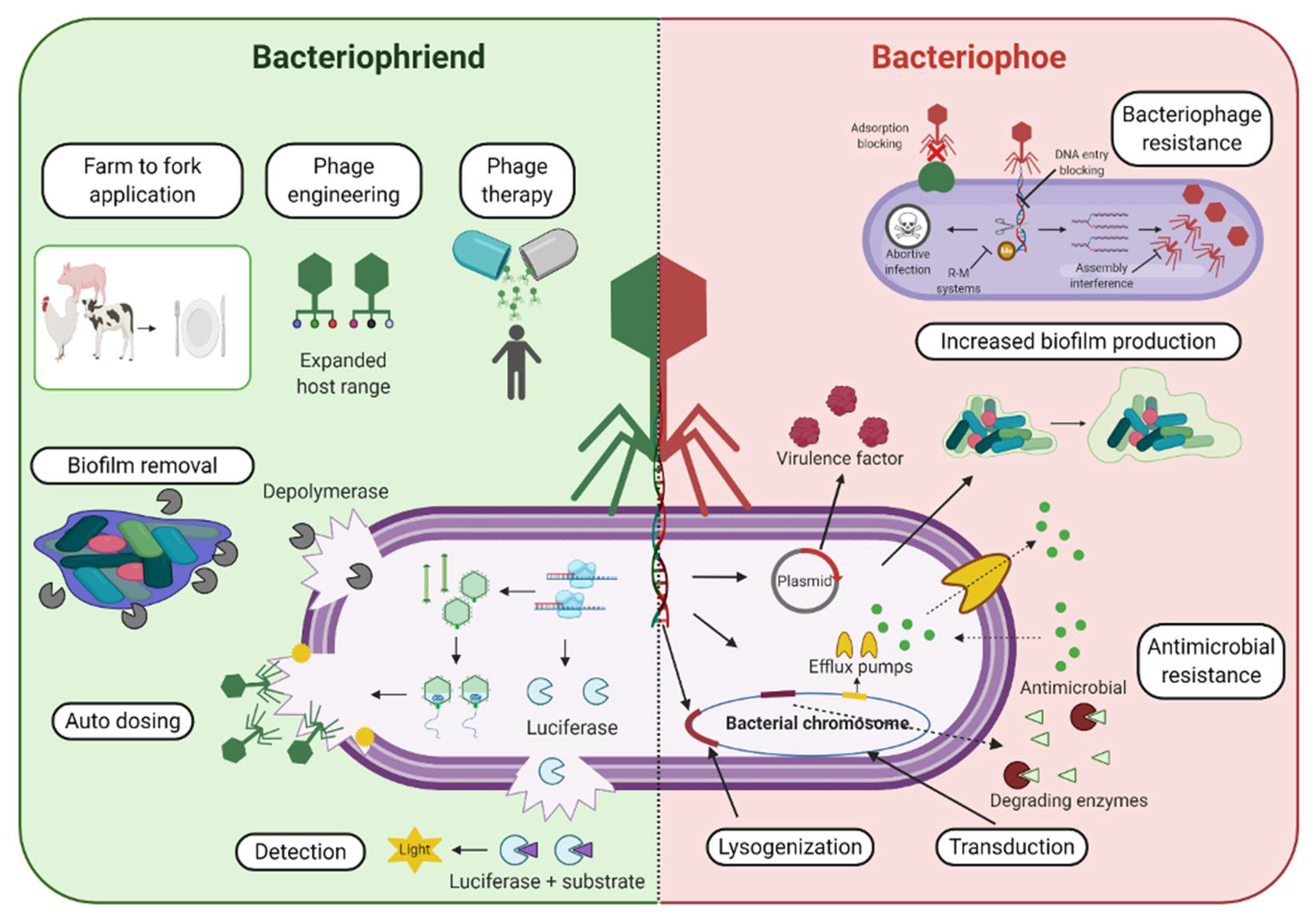
Therefore, we aim to provide a balanced perspective on bacteriophages (Figure 1).
Figure 1. Bacteriophages and the Antimicrobial Resistance Crisis: Friend or Foe? An overview of various phage-based applications and examples that display the potential benefit and “friend” aspect of bacteriophages (left, green background side) in combatting antimicrobial resistant bacterial strains, as well as some potentially detrimental or negatively associated outcomes that can contribute to the “foe” aspect of phage-based use (right, red background side) and their role in the acquisition, maintenance, and dissemination of antibacterial resistance genes.
2. Phage Selection Criteria for Biocontrol and Therapy
Bacteriophages must meet several parameters in order to ensure their safety and efficacy before they can be used for biocontrol and therapeutic purposes. Firstly, it is accepted that selected phages must be virulent (strictly lytic), thus lacking the ability to lysogenize targeted hosts [7]. This can help to minimize the transduction potential of the infecting bacteriophage, including the transfer of genes associated with resistance and virulence as discussed in the previous section. The lifestyle of a phage and the presence of potentially dangerous genetic determinants may be analyzed through the use of microbiological techniques by testing their ability to transfer selected markers between bacterial strains, through PCR-based techniques such as testing for the presence of bacterial DNA in phage particles, and through the use of genome sequencing and bioinformatic predictions in order to search for specific sequences associated with gene integration and toxin production [6]. Interestingly, advances in sequencing capabilities and synthetic biology techniques have led to innovative approaches incorporating the use of temperate phages and their lytic variants in phage therapy [8]. While strictly lytic phages will likely be the agent of choice in coming years, these advances highlight the potential viability and value of using temperate phages, including taking advantage of their natural ability for genome integration to either directly kill targeted bacteria through interference with host metabolism, or by rendering them as less pathogenic [8].
Moreover, since a high number of therapeutic phages ( at least 1 × 108 plaque-forming units (PFU)/mL) must be used in order to ensure sufficient contact and rapid infection of targeted cells [9], selected phages should be easily propagated in liquid media with high titer. The host range of selected phages must also be considered, with all epidemiologically important strains of a target bacterium being covered, as some bacterial strains contain several serovars which need to be managed [9]. An optimal balance between too narrow of a host range (which could lead to some strains of the same species not being affected) and too broad of a host range (which could lead to the killing of beneficial bacteria present) must be found [6]. While phages used for therapeutic purposes may follow a “personalized medicine” approach, where pathogenic strains are identified within an individual and specific phages are then selected from pre-existing banks for treatment [10], using phage cocktails, where phages of different specificities are utilized in therapeutic applications, is recommended [6]. The continuous arms race proceeding between bacteriophages and their bacterial hosts relates to the close relationship shared between bacteria and bacteriophages, where bacteria are under constant pressure from their viral invaders, and thus seek to gain resistance to phages to prolong their survival. While being the most abundant living organisms on the planet, bacteria are outnumbered by a factor of 10 to 1 by phages which infect them [11]. As such, they have evolved various phage resistance mechanisms including preventing phage adsorption, preventing phage DNA entry, cutting of phage nucleic acids, and abortive infection systems [12]. Therefore, the use of phage cocktails can aid in overcoming limitations associated with the use of monospecific bacteriophages, such as phage resistance development and limited host range profile [13][14]. Phage cocktails broaden the phage host range and improve treatment efficacy by increasing the number of targeted pathogens. Different phages in cocktails can also synergize by targeting different receptors on bacterial surfaces [15]. Phage cocktails have also been reported to be economically advantageous in comparison to the “personalized medicine” approach of other phage therapy treatments [15]. Improved bioengineering capabilities may also benefit the use of phage cocktails through expanded host ranges and greater utility and specificity in combination therapies with antibiotics, for example [13]. Moreover, the stability of selected phages under a variety of storage and application stages must be considered in order to ensure their durability within intended-use environments [9]. Numerous external factors may influence the integrity of bacteriophages and must be carefully considered when preparing phage preparations for therapeutic and biocontrol use, including temperature, acidity, and salinity or ion concentration [16]. Crucially, the varying levels of these parameters under physiological conditions (e.g., low stomach pH) or industrial settings (e.g., variable temperature) offers a challenge when selecting appropriate phages. As such, efforts to preserve, prolong, and optimize selected phages against the influence of these external factors include the utilization of adapted phage evolution, as well as phage formulation, stabilization, and encapsulation techniques [17][18][19][20]. For example, Kering et al. 2020 were able to induce improved thermal stability of phages at elevated temperatures without affecting their lytic activity [21]. Effective methods for the rapid enhancement of additional, desired characteristics among selected phages continue to be developed as well [22].
3. Bacteriophages for Detection of Bacterial Pathogens
In addition to their use as therapeutic agents against bacterial pathogens, bacteriophages also have the potential to be used for the rapid, specific, and sensitive detection of bacterial pathogens [23]. Their innate receptor specificity allows for the development of assays tailored to capture target bacteria, and their abundance in nature and ability to propagate within host cells provides increased sensitivity for assays using the “built-in” amplification system while also making them inexpensive and easy to produce [9]. While traditional culture-based detection remains the gold-standard for pathogen detection, these techniques are labor- and time-intensive, requiring 3–5 days for accurate results to be obtained [24]. Alternatively, bacteriophage-based detection allows for the more rapid detection of pathogens as the entire infection process only takes 1–2 h [9].
While the notion of using bacteriophages for the detection of bacterial pathogens has been examined throughout the past several decades, few examples of commercially available phage-based diagnostic tests have materialized. This can be attributed to the lack of sufficient knowledge on phage biology and genetic structure [9], in addition to the lack of protocols which ensure proper sensitivity, stability, and reproducibility required for phage-based detections methods to be successful [25]. Nonetheless, recent developments and extensive research have the potential to yield significant advantages for the improved sensitivity, specificity, and rapidity of phage-based detection methods [26]; paving the path for the increased impact of these methods and a move away from culture-based detection techniques [27]. Phage-based detection methods have been characterized in relation to their mechanism of action, with infection-based and capture-based detection methods being most prevalent [25], as briefly highlighted below.
3.1. Infection-Based Detection
3.2. Capture-Based Detection
Capture-based detection employs the unique specificity of bacteriophages to utilize them as biosensors [25]. Capture-based techniques may utilize whole-phage particles or specific phage receptors as tag molecules, using their innate affinity to detect targeted pathogens without the need for phage infection [26]. The immobilization of phage virions enables them to be used as specific bioreceptors upon the detection of their binding to specific bacterial hosts [23][25]. One of the most common methods employed to detect the binding of whole phage particles is surface plasmon resonance, which has been used to detect methicillin-resistant S. aureus (MRSA) at levels of 103 CFU/mL while also being able to distinguish methicillin-sensitive S. aureus from MRSA [37]. Conversely, the use of phage components as opposed to whole-phage particles offers a variety of benefits, including enhanced binding activity due to smaller probe sizes, improved specificity and affinity through engineering, and heightened robustness [23]. Specialized receptor binding proteins (RBPs) from tail fibers and spikes have been used in the glycotyping and identification of Salmonella strains [38] and Listeria strains [39]. In addition, genetically engineered tail-spike proteins from Salmonella phage P22 were used to create a biosensor that was able to detect real-time interactions of Salmonella cells at concentrations of 103 CFU/mL [40]. Receptor binding proteins have also been used in the detection of Campylobacter jejuni and Campylobacter coli, with produced assays exhibiting 100% specificity to both pathogens, and 95% sensitivity for C. jejuni and 90% sensitivity for C. coli [41]. Moreover, labelled cell wall binding domains (CBDs), sourced from phage endolysin enzymes, have also been used and proven effective in the capture and detection of various Gram-positive bacterial pathogens such as Listeria monocytogenes, Bacillus cereus, and Clostridium perfringens [26]. They have exhibited remarkable versatility, with their binding spectra being observed to be broader than the host ranges of corresponding phages [42], to displaying specificity at the serovar or strain level in the case of studied Listeria CBDs [43][44].
This entry is adapted from the peer-reviewed paper 10.3390/ph14030199
References
- Moghadam, M.T.; Amirmozafari, N.; Shariati, A.; Hallajzadeh, M.; Mirkalantari, S.; Khoshbayan, A.; Masjedian Jazi, F. How Phages Overcome the Challenges of Drug Resistant Bacteria in Clinical Infections. Infect. Drug Resist. 2020, 13, 45–61.
- Lin, D.M.; Koskella, B.; Lin, H.C. Phage therapy: An alternative to antibiotics in the age of multi-drug resistance. World J. Gastrointest. Pharmacol. Ther. 2017, 8, 162–173.
- Loc-Carrillo, C.; Abedon, S.T. Pros and cons of phage therapy. Bacteriophage 2011, 1, 111–114.
- Abedon, S.T.; Kuhl, S.J.; Blasdel, B.G.; Kutter, E.M. Phage treatment of human infections. Bacteriophage 2011, 1, 66–85.
- Fernández, L.; Gutiérrez, D.; Rodríguez, A.; García, P. Application of Bacteriophages in the Agro-Food Sector: A Long Way Toward Approval. Front. Cell. Infect. Microbiol. 2018, 8, 296.
- Reuter, M.; Kruger, D.H. Approaches to optimize therapeutic bacteriophage and bacteriophage-derived products to combat bacterial infections. Virus Genes 2020, 56, 136–149.
- Hyman, P. Phages for Phage Therapy: Isolation, Characterization, and Host Range Breadth. Pharmaceuticals 2019, 12, 35.
- Monteiro, R.; Pires, D.P.; Costa, A.R.; Azeredo, J. Phage Therapy: Going Temperate? Trends Microbiol. 2019, 27, 368–378.
- Brovko, L.Y.; Anany, H.; Griffiths, M.W. Chapter Six—Bacteriophages for Detection and Control of Bacterial Pathogens in Food and Food-Processing Environment; Henry, J.B.T.-A., Ed.; Academic Press: Cambridge, MA, USA, 2012; Volume 67, pp. 241–288. ISBN 1043-4526.
- Górski, A.; Borysowski, J.; Międzybrodzki, R. Phage Therapy: Towards a Successful Clinical Trial. Antibiotics 2020, 9, 827.
- Stern, A.; Sorek, R. The phage-host arms race: Shaping the evolution of microbes. Bioessays 2011, 33, 43–51.
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327.
- Altamirano, F.L.G.; Barr, J.J. Phage Therapy in the Postantibiotic Era. Clin. Microbiol. Rev. 2019, 32, e00066-18.
- Chan, B.K.; Abedon, S.T.; Loc-Carrillo, C. Phage cocktails and the future of phage therapy. Future Microbiol. 2013, 8, 769–783.
- Oechslin, F. Resistance Development to Bacteriophages Occurring during Bacteriophage Therapy. Viruses 2018, 10, 351.
- Jończyk, E.; Kłak, M.; Międzybrodzki, R.; Górski, A. The influence of external factors on bacteriophages—review. Folia Microbiol. 2011, 56, 191–200.
- Malik, D.J.; Sokolov, I.J.; Vinner, G.K.; Mancuso, F.; Cinquerrui, S.; Vladisavljevic, G.T.; Clokie, M.R.J.; Garton, N.J.; Stapley, A.G.F.; Kirpichnikova, A. Formulation, stabilisation and encapsulation of bacteriophage for phage therapy. Adv. Colloid Interface Sci. 2017, 249, 100–133.
- Petsong, K.; Benjakul, S.; Vongkamjan, K. Optimization of wall material for phage encapsulation via freeze-drying and antimicrobial efficacy of microencapsulated phage against Salmonella. J. Food Sci. Technol. 2020.
- Loh, B.; Gondil, V.S.; Manohar, P.; Khan, F.M.; Yang, H.; Leptihn, S. Encapsulation and Delivery of Therapeutic Phages. Appl. Environ. Microbiol. 2020, 87.
- Lone, A.; Anany, H.; Hakeem, M.; Aguis, L.; Avdjian, A.-C.; Bouget, M.; Atashi, A.; Brovko, L.; Rochefort, D.; Griffiths, M.W. Development of prototypes of bioactive packaging materials based on immobilized bacteriophages for control of growth of bacterial pathogens in foods. Int. J. Food Microbiol. 2016, 217, 49–58.
- Kering, K.K.; Zhang, X.; Nyaruaba, R.; Yu, J.; Wei, H. Application of adaptive evolution to improve the stability of bacteriophages during storage. Viruses 2020, 12, 423.
- Favor, A.H.; Llanos, C.D.; Youngblut, M.D.; Bardales, J.A. Optimizing bacteriophage engineering through an accelerated evolution platform. Sci. Rep. 2020, 10, 13981.
- Anany, H.; Chou, Y.; Cucic, S.; Derda, R.; Evoy, S.; Griffiths, M.W. From Bits and Pieces to Whole Phage to Nanomachines: Pathogen Detection Using Bacteriophages. Annu. Rev. Food Sci. Technol. 2017, 8, 305–329.
- Wisuthiphaet, N.; Yang, X.; Young, G.M.; Nitin, N. Rapid detection of Escherichia coli in beverages using genetically engineered bacteriophage T7. AMB Express 2019, 9, 55.
- Richter, Ł.; Janczuk-Richter, M.; Niedziółka-Jönsson, J.; Paczesny, J.; Hołyst, R. Recent advances in bacteriophage-based methods for bacteria detection. Drug Discov. Today 2018, 23, 448–455.
- Schmelcher, M.; Loessner, M.J. Application of bacteriophages for detection of foodborne pathogens. Bacteriophage 2014, 4, e28137.
- Schofield, D.; Sharp, N.J.; Westwater, C. Phage-based platforms for the clinical detection of human bacterial pathogens. Bacteriophage 2012, 2, 105–121.
- Schenborn, E.; Groskreutz, D. Reporter gene vectors and assays. Mol. Biotechnol. 1999, 13, 29–44.
- Meile, S.; Kilcher, S.; Loessner, M.J.; Dunne, M. Reporter Phage-Based Detection of Bacterial Pathogens: Design Guidelines and Recent Developments. Viruses 2020, 12, 944.
- Šuster, K.; Podgornik, A.; Cör, A. Quick bacteriophage-mediated bioluminescence assay for detecting Staphylococcus spp. in sonicate fluid of orthopaedic artificial joints. New Microbiol. 2017, 40, 190–196.
- Meile, S.; Sarbach, A.; Du, J.; Schuppler, M.; Saez, C.; Loessner, M.J.; Kilcher, S. Engineered Reporter Phages for Rapid Bioluminescence-Based Detection and Differentiation of Viable Listeria Cells. Appl. Environ. Microbiol. 2020, 86, e00442-20.
- Kim, S.; Kim, M.; Ryu, S. Development of an Engineered Bioluminescent Reporter Phage for the Sensitive Detection of Viable Salmonella Typhimurium. Anal. Chem. 2014, 86, 5858–5864.
- Ripp, S.; Jegier, P.; Johnson, C.M.; Brigati, J.R.; Sayler, G.S. Bacteriophage-amplified bioluminescent sensing of Escherichia coli O157:H7. Anal. Bioanal. Chem. 2008, 391, 507–514.
- Vinay, M.; Franche, N.; Grégori, G.; Fantino, J.-R.; Pouillot, F.; Ansaldi, M. Phage-Based Fluorescent Biosensor Prototypes to Specifically Detect Enteric Bacteria Such as E. coli and Salmonella enterica Typhimurium. PLoS ONE 2015, 10, e0131466.
- Chang, T.C.; Ding, H.C.; Chen, S. A Conductance Method for the Identification of Escherichia coli O157:H7 Using Bacteriophage AR1. J. Food Prot. 2002, 65, 12–17.
- Swift, B.M.C.; Meade, N.; Barron, E.S.; Bennett, M.; Perehenic, T.; Hughes, V.; Stevenson, K.; Rees, C.E.D. The development and use of Actiphage® to detect viable mycobacteria from bovine tuberculosis and Johne’s disease-infected animals. Microb. Biotechnol. 2020, 13, 738–746.
- Tawil, N.; Sacher, E.; Mandeville, R.; Meunier, M. Surface plasmon resonance detection of E. coli and methicillin-resistant S. aureus using bacteriophages. Biosens. Bioelectron. 2012, 37, 24–29.
- Schmidt, A.; Rabsch, W.; Broeker, N.K.; Barbirz, S. Bacteriophage tailspike protein based assay to monitor phase variable glucosylations in Salmonella O-antigens. BMC Microbiol. 2016, 16, 207.
- Sumrall, E.T.; Röhrig, C.; Hupfeld, M.; Selvakumar, L.; Du, J.; Dunne, M.; Schmelcher, M.; Shen, Y.; Loessner, M.J. Glycotyping and Specific Separation of Listeria monocytogenes with a Novel Bacteriophage Protein Tool Kit. Appl. Environ. Microbiol. 2020, 86, e00612-20.
- Singh, A.; Arya, S.K.; Glass, N.; Hanifi-Moghaddam, P.; Naidoo, R.; Szymanski, C.M.; Tanha, J.; Evoy, S. Bacteriophage tailspike proteins as molecular probes for sensitive and selective bacterial detection. Biosens. Bioelectron. 2010, 26, 131–138.
- Javed, M.A.; Poshtiban, S.; Arutyunov, D.; Evoy, S.; Szymanski, C.M. Bacteriophage Receptor Binding Protein Based Assays for the Simultaneous Detection of Campylobacter jejuni and Campylobacter coli. PLoS ONE 2013, 8, e69770.
- Gu, J.; Lu, R.; Liu, X.; Han, W.; Lei, L.; Gao, Y.; Zhao, H.; Li, Y.; Diao, Y. LysGH15B, the SH3b Domain of Staphylococcal Phage Endolysin LysGH15, Retains High Affinity to Staphylococci. Curr. Microbiol. 2011, 63, 538.
- Loessner, M.J.; Kramer, K.; Ebel, F.; Scherer, S. C-terminal domains of Listeria monocytogenes bacteriophage murein hydrolases determine specific recognition and high-affinity binding to bacterial cell wall carbohydrates. Mol. Microbiol. 2002, 44, 335–349.
- Kretzer, J.W.; Schmelcher, M.; Loessner, M.J. Ultrasensitive and Fast Diagnostics of Viable Listeria Cells by CBD Magnetic Separation Combined with A511::luxAB Detection. Viruses 2018, 10, 626.