Circulating tumour cells (CTCs) are the precursor cells for the formation of metastatic disease.
- circulating tumour cell (CTC)
- cancer
- metastasis
- liquid biopsy
1. Background
With a simple blood draw, liquid biopsies enable the non-invasive sampling of CTCs from the blood, which have the potential to provide important insights into cancer detection and monitoring. Since gaining FDA approval in 2004, the CellSearch system has been used to determine the prognosis of patients with metastatic breast, prostate and colorectal cancers. This utilises the cell surface marker Epithelial Cell Adhesion Molecule (EpCAM), to enrich CTCs, and many other technologies have adopted this approach. More recently, the role of mesenchymal-like CTCs in metastasis formation has come to light. It has been suggested that these cells are more aggressive metastatic precursors than their epithelial counterparts; however, mesenchymal CTCs remain undetected by EpCAM-based enrichment methods. This has prompted the development of a variety of ‘label free’ enrichment technologies, which exploit the unique physical properties of CTCs (such as size and deformability) compared to other blood components.
2. Introduction
Circulating tumour cells (CTCs) are shed into the bloodstream from both primary and metastatic tumours and those that are able to survive in the circulation represent metastatic precursor cells [1]. CTCs are important biomarkers for disease and are a powerful tool to study tumour progression and evolution. They represent a rare and heterogeneous population of cells, typically accounting for ∼1 cells for every 105–106 peripheral blood mononuclear cells (PBMCs), so a key challenge for their clinical utility is the development of standardised isolation and characterisation technologies [2]. There are numerous technologies that have been developed to enrich CTCs from normal hematopoietic cells that rely on physical and biological properties of CTCs, including size, density, cellular charge and expression of cellular markers. The enrichment techniques (Table 1) can broadly be divided into immunocapture methods that differentiate cells based on epithelial cell surface marker expression, notably epithelial cell adhesion molecule (EpCAM) (Figure 1A), and those that differentiate based on distinct biophysical properties (Figure 1B,C). If CTC enrichment and characterisation is to be routinely used in the clinical setting, technologies must ideally meet several criteria: they must have high detection and recovery rates, with accurate throughput sample processing and enumeration capability. Further, they must be generally fully automated and easy to use, with little to no pre-processing of blood required. Finally, if they are to have wide clinical applicability, they must be able to detect heterogeneous cells from a wide range of different cancers.
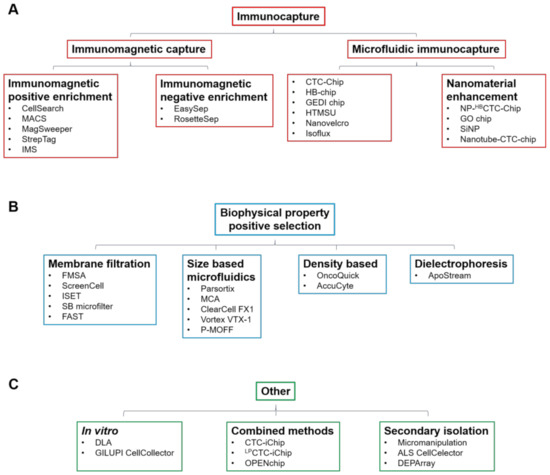
Table 1. CTC isolation technologies, grouped based on enrichment method. Capture efficiency, recovery rate and advantages and disadvantages of the technologies are also shown.
| Subcategory | Name | Capture Efficiency (%) | Recovery Rate (%) | Advantages | Disadvantages |
|---|---|---|---|---|---|
| Immunomagnetic enrichment | |||||
| Immunomagnetic positive enrichment | CellSearch [3][4][5][6][7] | 42–90 | Semi automated Can process up to 8 samples at a time In device staining CTC enumeration via CellTracks Analyser FDA approved |
Recovery of EpCAM+ CTCs only Only able to detect CTCs expressing high levels of EpCAM |
|
| MACS [8][9] | 25–90 | Cocktail of antibodies available to increase CTC capture Able to process up to 15 mL blood Easy elution of CTCs Pro Separator can process up to 6 samples at once |
Recovery of EpCAM+ CTCs only Suggested the MACS system is better suited for tissue samples |
||
| MagSweeper [10][11] | 60–70 | Nonadherent plastic sleeves allow for multiple rounds of capture to increase capture efficiency | Recovery of EpCAM+ CTCs only | ||
| Strep-tag [12][13] | 79–86 | 70 | Easy release of CTCs by simple addition of d-biotin Possibility to use a cocktail of antibodies to increase capture |
Recovery of EpCAM+ CTCs only | |
| IMS [14] | 92 | Leukocytes repelled so high purity recoveries | Recovery of EpCAM+ CTCs only Not yet tested on patient samples |
||
| Immunomagnetic negative enrichment | EasySep [15][16] | 19–65 | Recovery of heterogeneous population of CTCs | Exclusion of CTC-WBC clusters Variable recovery rates May inadvertantly remove CTCs |
|
| RosetteSep [17] | 62.5 | Recovery of heterogeneous population of CTCs Cocktail of antibodies used to maximise depletion |
Exclusion of CTC-WBC clusters, May inadvertantly remove CTCs |
||
| Microfluidic immunocapture positive enrichment | CTC-Chip [18][19] | >60 | Large surface area for CTC capture High viability of recovered cells |
Recovery of EpCAM+ CTCs only Slow processing rate Complex geometry of chip difficult to scale up Geometry prevents passage of CTC clusters |
|
| HB-chip [20] | 74.5–97 | HB grooves increase CTC-antibody contact for increased cell capture | Recovery of EpCAM+ CTCs only | ||
| GEDI chip [21] | 80–90 | Large surface area for CTC capture Possibility to functionalise with alternative antibodies |
May miss heterogeneity of CTCs | ||
| HTMSU [22] | >97 | Quick processing On-chip single-cell conductometric counting for enumeration |
Recovery of EpCAM+ CTCs only | ||
| Nanovelcro [23] | 70–95 | 4 generations developed for different clinical utilities 3rd and 4th generation chips adapted for easy CTC release |
Recovery of EpCAM+ CTCs only | ||
| Isoflux [4] | 74–90 | 64–75 | Utilises microfluidic approach to increase EpCAM sensitivity Up to 4 samples can be processed in parallel Multiple kits including cocktails of antibodies to capture heterogeneity IsoFlux Cytation Imager for sample scanning |
||
| Capture enhancement by nanomaterials | NP-HBCTC-Chip [24] | 79–97 | Simple release of CTCs by addition of glutathione (GSH) Chip surface can be functionalised with a cocktail of antibodies for enhanced capture efficiency |
Recovery of EpCAM+ CTCs only Very low throughput |
|
| GO chip [25][26] | 67–100 | 91–95 | Simple chip design Large surface area for increased CTC capture |
Recovery of EpCAM+ CTCs only | |
| SiNP [27] | 84–91 | Large surface area for CTC capture | Recovery of EpCAM+ CTCs only | ||
| Capture enhancement by nanomaterials | Nanotube-CTC-chip [28] | 89–100 | Preferential adherence negates need for EpCAM antibodies Planar enrichment surface makes chip visualisation and imaging easy |
Time taken for optimal CTC adherence to substrate is too long | |
| Size based enrichment | |||||
| Membrane filtration | FMSA [29] | 90 | Recovery of heterogeneous population of CTCs Cheap and easy to produce Quick processing time |
Filter clogging highly likely | |
| ScreenCell [30] | 74–91 | Recovery of heterogeneous population of CTCs Cheap and easy to produce Three different devices offered depending on downstream requirements Quick processing time |
Unevenly distributed or fused pores can reduce capture efficiency | ||
| ISET [31][32] | 83–100 | Recovery of heterogeneous population of CTCs Cheap and easy to produce Ability to process 12 samples in parallel |
Slow processing time Blood must be diluted 1:10 to prevent membrane clogging |
||
| SB microfilter [33] | 78–83 | Recovery of heterogeneous population of CTCs Cheap and easy to produce Quick processing time |
Only 1 mL blood can be processed at a time due to device clogging | ||
| FAST [34] | 94–98 | Recovery of heterogeneous population of CTCs Cheap and easy to produce Quick processing time |
|||
| Microfluidics | Parsortix [35] | 42–70 | 54–69 | Recovery of heterogeneous population of CTCs Ability to capture CTC clusters Option for on-chip staining |
Slow processing time On-chip imaging difficult |
| MCA [36] | >90 | 68–100 | Recovery of heterogeneous population of CTCs Option for on-chip staining Ability to process up to 4 samples in parallel |
||
| ClearCell FX1 [37][38] | 52–79 | Recovery of heterogeneous population of CTCs Quick processing time No channel clogging observed |
|||
| Vortex VTX-1 [39][40] | 53.8–71.6 | Recovery of heterogeneous population of CTCs Filters at channel inlet prevent channel clogging Fully automated process Quick processing time Associated BioView for enumeration Option to run in “high recovery” or “high purity” mode |
|||
| p-MOFF [41] | 91.6–93.75 | Recovery of heterogeneous population of CTCs Quick processing time No channel clogging observed |
RBC lysis and Ficoll density centrifugation required | ||
| Density based | OncoQuick [42][43] | 25–87 | Recovery of heterogeneous population of CTCs Up to 25 mL blood can be processed per tube |
Low detection and recoveryrates | |
| AccuCyte [44] | 81–90.5 | Recovery of heterogeneous population of CTCs Allows for processing of multiple samples in parallel Associated CyteFinder and CytePicker systems for imaging and mechanical selection of CTCs |
|||
| Other | |||||
| Dielectrophoresis | ApoStream [45][46] | 55–78.5 | Recovery of heterogeneous population of CTCs Quick processing time iCys laser scanning cytometer for enumeration High viability of recovered cells |
||
| In vivo | Diagnostic leukapheresis (DLA) [47] | Recovery of heterogeneous population of CTCs Recovery of much greater numbers of CTCs |
Only a pre-enrichment step so must be used in combination with another enrichment technology Huge leukocyte background |
||
| GILUPI CellCollector [48] | Potential for much greater numbers recovered | More invasive for the patient than a simple blood draw Recovery of EpCAM+ CTCs only |
|||
| Combined | CTC-iChip [49] | 70–100 | Option for positive or negative enrichment approach Inertial focusing provides high sensitivity selection Quick processing time |
Positive enrichment only allows for recovery of EpCAM+ CTCs Negative enrichment will exclude CTC-WBC clusters |
|
| LPCTC-iChip [50] | 85.5–100 | Potential for much greater numbers recovered Magnetic field directs WBCs to centre of channel to prevent channel clogging Extremely high throughput |
Disregards CTC-WBC clusters Initial debulking step may result in CTC loss |
||
| OPENchip [51] | 50 | Chip allows for CTC enrichment and on-chip downstream molecular analysis | Low throughput, low recovery rates |
This entry is adapted from the peer-reviewed paper 10.3390/cancers13050970
References
- Yu, M.; Stott, S.; Toner, M.; Maheswaran, S.; Haber, D.A. Circulating tumor cells: Approaches to isolation and characterization. J. Cell Biol. 2011, 192, 373–382.
- Ross, A.A.; Cooper, B.W.; Lazarus, H.M.; Mackay, W.; Moss, T.J.; Ciobanu, N.; Tallman, M.S.; Kennedy, M.J.; Davidson, N.E.; Sweet, D.; et al. Detection and viability of tumor cells in peripheral blood stem cell collections from breast cancer patients using immunocytochemical and clonogenic assay techniques. Blood 1993, 82, 2605–2610.
- Punnoose, E.A.; Atwal, S.K.; Spoerke, J.M.; Savage, H.; Pandita, A.; Yeh, R.F.; Pirzkall, A.; Fine, B.M.; Amler, L.C.; Chen, D.S.; et al. Molecular biomarker analyses using circulating tumor cells. PLoS ONE 2010, 5, e12517.
- Harb, W.; Fan, A.; Tran, T.; Danila, D.C.; Keys, D.; Schwartz, M.; Ionescu-Zanetti, C. Mutational Analysis of Circulating Tumor Cells Using a Novel Microfluidic Collection Device and qPCR Assay. Transl. Oncol. 2013, 6, 528–538.
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791.
- Politaki, E.; Agelaki, S.; Apostolaki, S.; Hatzidaki, D.; Strati, A.; Koinis, F.; Perraki, M.; Saloustrou, G.; Stoupis, G.; Kallergi, G.; et al. A Comparison of Three Methods for the Detection of Circulating Tumor Cells in Patients with Early and Metastatic Breast Cancer. Cell. Physiol. Biochem. 2017, 44, 594–606.
- Mikolajczyk, S.D.; Millar, L.S.; Tsinberg, P.; Coutts, S.M.; Zomorrodi, M.; Pham, T.; Bischoff, F.Z.; Pircher, T.J. Detection of EpCAM-Negative and Cytokeratin-Negative Circulating Tumor Cells in Peripheral Blood. J. Oncol. 2011, 2011, 252361.
- Miltenyi, S.; Müller, W.; Weichel, W.; Radbruch, A. High gradient magnetic cell separation with MACS. Cytometry 1990, 11, 231–238.
- Zhu, D.M.; Wu, L.; Suo, M.; Gao, S.; Xie, W.; Zan, M.H.; Liu, A.; Chen, B.; Wu, W.T.; Ji, L.W.; et al. Engineered red blood cells for capturing circulating tumor cells with high performance. Nanoscale 2018, 10, 6014–6023.
- Talasaz, A.H.; Powell, A.A.; Huber, D.E.; Berbee, J.G.; Roh, K.-H.; Yu, W.; Xiao, W.; Davis, M.M.; Pease, R.F.; Mindrinos, M.N.; et al. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proc. Natl. Acad. Sci. USA 2009, 106, 3970–3975.
- Cann, G.M.; Gulzar, Z.G.; Cooper, S.; Li, R.; Luo, S.; Tat, M.; Stuart, S.; Schroth, G.; Srinivas, S.; Ronaghi, M.; et al. mRNA-Seq of single prostate cancer circulating tumor cells reveals recapitulation of gene expression and pathways found in prostate cancer. PLoS ONE 2012, 7, e49144.
- Schmidt, T.G.; Skerra, A. The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat. Protoc. 2007, 2, 1528–1535.
- Lu, N.-N.; Xie, M.; Wang, J.; Lv, S.-W.; Yi, J.-S.; Dong, W.-G.; Huang, W.-H. Biotin-Triggered Decomposable Immunomagnetic Beads for Capture and Release of Circulating Tumor Cells. ACS Appl. Mater. Interfaces 2015, 7, 8817–8826.
- Xiong, K.; Wei, W.; Jin, Y.; Wang, S.; Zhao, D.; Wang, S.; Gao, X.; Qiao, C.; Yue, H.; Ma, G.; et al. Biomimetic Immuno-Magnetosomes for High-Performance Enrichment of Circulating Tumor Cells. Adv. Mater. 2016, 28, 7929–7935.
- Drucker, A.; Teh, E.M.; Kostyleva, R.; Rayson, D.; Douglas, S.; Pinto, D.M. Comparative performance of different methods for circulating tumor cell enrichment in metastatic breast cancer patients. PLoS ONE 2020, 15, e0237308.
- Liu, Z.; Fusi, A.; Klopocki, E.; Schmittel, A.; Tinhofer, I.; Nonnenmacher, A.; Keilholz, U. Negative enrichment by immunomagnetic nanobeads for unbiased characterization of circulating tumor cells from peripheral blood of cancer patients. J. Transl. Med. 2011, 9, 70.
- He, W.; Kularatne, S.A.; Kalli, K.R.; Prendergast, F.G.; Amato, R.J.; Klee, G.G.; Hartmann, L.C.; Low, P.S. Quantitation of circulating tumor cells in blood samples from ovarian and prostate cancer patients using tumor-specific fluorescent ligands. Int. J. Cancer 2008, 123, 1968–1973.
- Sequist, L.V.; Nagrath, S.; Toner, M.; Haber, D.A.; Lynch, T.J. The CTC-chip: An exciting new tool to detect circulating tumor cells in lung cancer patients. J. Thorac. Oncol. 2009, 4, 281–283.
- Nagrath, S.; Sequist, L.V.; Maheswaran, S.; Bell, D.W.; Irimia, D.; Ulkus, L.; Smith, M.R.; Kwak, E.L.; Digumarthy, S.; Muzikansky, A.; et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007, 450, 1235–1239.
- Stott, S.L.; Hsu, C.H.; Tsukrov, D.I.; Yu, M.; Miyamoto, D.T.; Waltman, B.A.; Rothenberg, S.M.; Shah, A.M.; Smas, M.E.; Korir, G.K.; et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc. Natl. Acad. Sci. USA 2010, 107, 18392–18397.
- Gleghorn, J.P.; Pratt, E.D.; Denning, D.; Liu, H.; Bander, N.H.; Tagawa, S.T.; Nanus, D.M.; Giannakakou, P.A.; Kirby, B.J. Capture of circulating tumor cells from whole blood of prostate cancer patients using geometrically enhanced differential immunocapture (GEDI) and a prostate-specific antibody. Lab Chip 2010, 10, 27–29.
- Adams, A.A.; Okagbare, P.I.; Feng, J.; Hupert, M.L.; Patterson, D.; Göttert, J.; McCarley, R.L.; Nikitopoulos, D.; Murphy, M.C.; Soper, S.A. Highly efficient circulating tumor cell isolation from whole blood and label-free enumeration using polymer-based microfluidics with an integrated conductivity sensor. J. Am. Chem. Soc. 2008, 130, 8633–8641.
- Jan, Y.J.; Chen, J.-F.; Zhu, Y.; Lu, Y.-T.; Chen, S.H.; Chung, H.; Smalley, M.; Huang, Y.-W.; Dong, J.; Chen, L.-C.; et al. NanoVelcro rare-cell assays for detection and characterization of circulating tumor cells. Adv. Drug Deliv. Rev. 2018, 125, 78–93.
- Park, M.-H.; Reátegui, E.; Li, W.; Tessier, S.N.; Wong, K.H.K.; Jensen, A.E.; Thapar, V.; Ting, D.; Toner, M.; Stott, S.L.; et al. Enhanced Isolation and Release of Circulating Tumor Cells Using Nanoparticle Binding and Ligand Exchange in a Microfluidic Chip. J. Am. Chem. Soc. 2017, 139, 2741–2749.
- Yoon, H.J.; Kim, T.H.; Zhang, Z.; Azizi, E.; Pham, T.M.; Paoletti, C.; Lin, J.; Ramnath, N.; Wicha, M.S.; Hayes, D.F.; et al. Sensitive capture of circulating tumour cells by functionalized graphene oxide nanosheets. Nat. Nanotechnol. 2013, 8, 735–741.
- Yoon, H.J.; Shanker, A.; Wang, Y.; Kozminsky, M.; Jin, Q.; Palanisamy, N.; Burness, M.L.; Azizi, E.; Simeone, D.M.; Wicha, M.S.; et al. Tunable Thermal-Sensitive Polymer–Graphene Oxide Composite for Efficient Capture and Release of Viable Circulating Tumor Cells. Adv. Mater. 2016, 28, 4891–4897.
- Wang, S.; Wang, H.; Jiao, J.; Chen, K.-J.; Owens, G.E.; Kamei, K.-i.; Sun, J.; Sherman, D.J.; Behrenbruch, C.P.; Wu, H.; et al. Three-Dimensional Nanostructured Substrates toward Efficient Capture of Circulating Tumor Cells. Angew. Chem. Int. Ed. 2009, 48, 8970–8973.
- Loeian, M.S.; Mehdi Aghaei, S.; Farhadi, F.; Rai, V.; Yang, H.W.; Johnson, M.D.; Aqil, F.; Mandadi, M.; Rai, S.N.; Panchapakesan, B. Liquid biopsy using the nanotube-CTC-chip: Capture of invasive CTCs with high purity using preferential adherence in breast cancer patients. Lab Chip 2019, 19, 1899–1915.
- Harouaka, R.A.; Zhou, M.-D.; Yeh, Y.-T.; Khan, W.J.; Das, A.; Liu, X.; Christ, C.C.; Dicker, D.T.; Baney, T.S.; Kaifi, J.T.; et al. Flexible Micro Spring Array Device for High-Throughput Enrichment of Viable Circulating Tumor Cells. Clin. Chem. 2014, 60, 323–333.
- Desitter, I.; Guerrouahen, B.S.; Benali-Furet, N.; Wechsler, J.; Jänne, P.A.; Kuang, Y.; Yanagita, M.; Wang, L.; Berkowitz, J.A.; Distel, R.J.; et al. A new device for rapid isolation by size and characterization of rare circulating tumor cells. Anticancer Res. 2011, 31, 427–441.
- Farace, F.; Massard, C.; Vimond, N.; Drusch, F.; Jacques, N.; Billiot, F.; Laplanche, A.; Chauchereau, A.; Lacroix, L.; Planchard, D.; et al. A direct comparison of CellSearch and ISET for circulating tumour-cell detection in patients with metastatic carcinomas. Br. J. Cancer 2011, 105, 847–853.
- Kallergi, G.; Politaki, E.; Alkahtani, S.; Stournaras, C.; Georgoulias, V. Evaluation of Isolation Methods for Circulating Tumor Cells (CTCs). Cell. Physiol. Biochem. 2016, 40, 411–419.
- Zhou, M.-D.; Hao, S.; Williams, A.J.; Harouaka, R.A.; Schrand, B.; Rawal, S.; Ao, Z.; Brenneman, R.; Gilboa, E.; Lu, B.; et al. Separable Bilayer Microfiltration Device for Viable Label-free Enrichment of Circulating Tumour Cells. Sci. Rep. 2014, 4, 7392.
- Kim, T.-H.; Lim, M.; Park, J.; Oh, J.M.; Kim, H.; Jeong, H.; Lee, S.J.; Park, H.C.; Jung, S.; Kim, B.C.; et al. FAST: Size-Selective, Clog-Free Isolation of Rare Cancer Cells from Whole Blood at a Liquid–Liquid Interface. Anal. Chem. 2017, 89, 1155–1162.
- Cohen, E.N.; Jayachandran, G.; Hardy, M.R.; Venkata Subramanian, A.M.; Meng, X.; Reuben, J.M. Antigen-agnostic microfluidics-based circulating tumor cell enrichment and downstream molecular characterization. PLoS ONE 2020, 15, e0241123.
- Hosokawa, M.; Hayata, T.; Fukuda, Y.; Arakaki, A.; Yoshino, T.; Tanaka, T.; Matsunaga, T. Size-Selective Microcavity Array for Rapid and Efficient Detection of Circulating Tumor Cells. Anal. Chem. 2010, 82, 6629–6635.
- Lee, Y.; Guan, G.; Bhagat, A.A. ClearCell(R) FX, a label-free microfluidics technology for enrichment of viable circulating tumor cells. Cytom. Part A 2018, 93, 1251–1254.
- Yap, Y.-S.; Leong, M.C.; Chua, Y.W.; Loh, K.W.J.; Lee, G.E.; Lim, E.H.; Dent, R.; Ng, R.C.H.; Lim, J.H.-C.; Singh, G.; et al. Detection and prognostic relevance of circulating tumour cells (CTCs) in Asian breast cancers using a label-free microfluidic platform. PLoS ONE 2019, 14, e0221305.
- Lemaire, C.A.; Liu, S.Z.; Wilkerson, C.L.; Ramani, V.C.; Barzanian, N.A.; Huang, K.-W.; Che, J.; Chiu, M.W.; Vuppalapaty, M.; Dimmick, A.M.; et al. Fast and Label-Free Isolation of Circulating Tumor Cells from Blood: From a Research Microfluidic Platform to an Automated Fluidic Instrument, VTX-1 Liquid Biopsy System. SLAS Technol. Transl. Life Sci. Innov. 2018, 23, 16–29.
- Sollier-Christen, E.; Renier, C.; Kaplan, T.; Kfir, E.; Crouse, S.C. VTX-1 Liquid Biopsy System for Fully-Automated and Label-Free Isolation of Circulating Tumor Cells with Automated Enumeration by BioView Platform. Cytom. Part A 2018, 93, 1240–1245.
- Hyun, K.A.; Kwon, K.; Han, H.; Kim, S.I.; Jung, H.I. Microfluidic flow fractionation device for label-free isolation of circulating tumor cells (CTCs) from breast cancer patients. Biosens. Bioelectron. 2013, 40, 206–212.
- Königsberg, R.; Obermayr, E.; Bises, G.; Pfeiler, G.; Gneist, M.; Wrba, F.; de Santis, M.; Zeillinger, R.; Hudec, M.; Dittrich, C. Detection of EpCAM positive and negative circulating tumor cells in metastatic breast cancer patients. Acta Oncol. 2011, 50, 700–710.
- Rosenberg, R.; Gertler, R.; Friederichs, J.; Fuehrer, K.; Dahm, M.; Phelps, R.; Thorban, S.; Nekarda, H.; Siewert, J.R. Comparison of two density gradient centrifugation systems for the enrichment of disseminated tumor cells in blood. Cytometry 2002, 49, 150–158.
- Campton, D.E.; Ramirez, A.B.; Nordberg, J.J.; Drovetto, N.; Clein, A.C.; Varshavskaya, P.; Friemel, B.H.; Quarre, S.; Breman, A.; Dorschner, M.; et al. High-recovery visual identification and single-cell retrieval of circulating tumor cells for genomic analysis using a dual-technology platform integrated with automated immunofluorescence staining. BMC Cancer 2015, 15, 360.
- Rugo, H.S.; Cortes, J.; Awada, A.; O’Shaughnessy, J.; Twelves, C.; Im, S.A.; Hannah, A.; Lu, L.; Sy, S.; Caygill, K.; et al. Change in Topoisomerase 1-Positive Circulating Tumor Cells Affects Overall Survival in Patients with Advanced Breast Cancer after Treatment with Etirinotecan Pegol. Clin. Cancer Res. 2018, 24, 3348–3357.
- Gupta, V.; Jafferji, I.; Garza, M.; Melnikova, V.O.; Hasegawa, D.K.; Pethig, R.; Davis, D.W. ApoStream(™), a new dielectrophoretic device for antibody independent isolation and recovery of viable cancer cells from blood. Biomicrofluidics 2012, 6, 24133.
- Fehm, T.N.; Meier-Stiegen, F.; Driemel, C.; Jäger, B.; Reinhardt, F.; Naskou, J.; Franken, A.; Neubauer, H.; Neves, R.P.L.; van Dalum, G.; et al. Diagnostic leukapheresis for CTC analysis in breast cancer patients: CTC frequency, clinical experiences and recommendations for standardized reporting. Cytom. Part A 2018, 93, 1213–1219.
- Saucedo-Zeni, N.; Mewes, S.; Niestroj, R.; Gasiorowski, L.; Murawa, D.; Nowaczyk, P.; Tomasi, T.; Weber, E.; Dworacki, G.; Morgenthaler, N.G.; et al. A novel method for the in vivo isolation of circulating tumor cells from peripheral blood of cancer patients using a functionalized and structured medical wire. Int. J. Oncol. 2012, 41, 1241–1250.
- Ozkumur, E.; Shah, A.M.; Ciciliano, J.C.; Emmink, B.L.; Miyamoto, D.T.; Brachtel, E.; Yu, M.; Chen, P.-I.; Morgan, B.; Trautwein, J.; et al. Inertial Focusing for Tumor Antigen–Dependent and –Independent Sorting of Rare Circulating Tumor Cells. Sci. Transl. Med. 2013, 5, 179ra147.
- Mishra, A.; Dubash, T.D.; Edd, J.F.; Jewett, M.K.; Garre, S.G.; Karabacak, N.M.; Rabe, D.C.; Mutlu, B.R.; Walsh, J.R.; Kapur, R.; et al. Ultrahigh-throughput magnetic sorting of large blood volumes for epitope-agnostic isolation of circulating tumor cells. Proc. Natl. Acad. Sci. USA 2020, 117, 16839–16847.
- Lee, A.C.; Svedlund, J.; Darai, E.; Lee, Y.; Lee, D.; Lee, H.-B.; Kim, S.-M.; Kim, O.; Bae, H.J.; Choi, A.; et al. OPENchip: An on-chip in situ molecular profiling platform for gene expression analysis and oncogenic mutation detection in single circulating tumour cells. Lab Chip 2020, 20, 912–922.
