Zinc finger myeloid, nervy, and deformed epidermal autoregulatory factor 1-type containing 8 (Zinc finger MYND-type containing 8, ZMYND8) is a transcription factor, a histone H3-interacting protein, and a putative chromatin reader/effector that plays an essential role in regulating transcription during normal cellular growth. Mutations and altered expression of ZMYND8 are associated with the development and progression of cancer. Increased expression of ZMYND8 is linked to breast, prostate, colorectal, and cervical cancers. It exerts pro-oncogenic effects in breast and prostate cancers, and it promotes angiogenesis in zebrafish, as well as in breast and prostate cancers. In contrast, downregulation of ZMYND8 is also reported in breast, prostate, and nasopharyngeal cancers. ZMYND8 acts as a tumor suppressor in breast and prostate cancers, and it inhibits tumor growth by promoting differentiation; inhibiting proliferation, cell-cycle progression, invasiveness, and metastasis; and maintaining the epithelial phenotype in various types of cancers. These data together suggest that ZMYND8 is important in tumorigenesis; however, the existing data are contradictory. More studies are necessary to clarify the exact role of ZMYND8 in tumorigenesis. In the future, regulation of expression/activity of ZMYND8 and/or its binding partners may become useful in treating cancer.
- ZMYND8
- tumorigenesis
- tumor suppression
1. Introduction
Zinc finger myeloid, nervy, and deformed epidermal autoregulatory factor 1-type containing 8 (Zinc finger MYND-type containing 8, ZMYND8) is a multifunctional transcription factor harboring conserved chromatin-binding module with affinity for chromatin [1]. It was initially identified as activated protein-kinase-C (PKC)-binding protein and is a member of the receptor for activated C-kinase (RACK) family proteins that anchor activated PKC and increase its phosphorylation and duration of inactivation; it is also called RACK7 [2][3]. ZMYND8 contains a Pro-Trp-Trp-Pro (PWWP) chromatin-binding domain, a bromodomain (BRD), a plant homeodomain (PHD) type zinc finger, and a MYND domain for protein–protein interaction (Figure 1) [3][4]. The PHD–BRD–PWWP (PBP) domains are histone readers, and the proteins containing PBP domains have various chromatin-related functions [5].
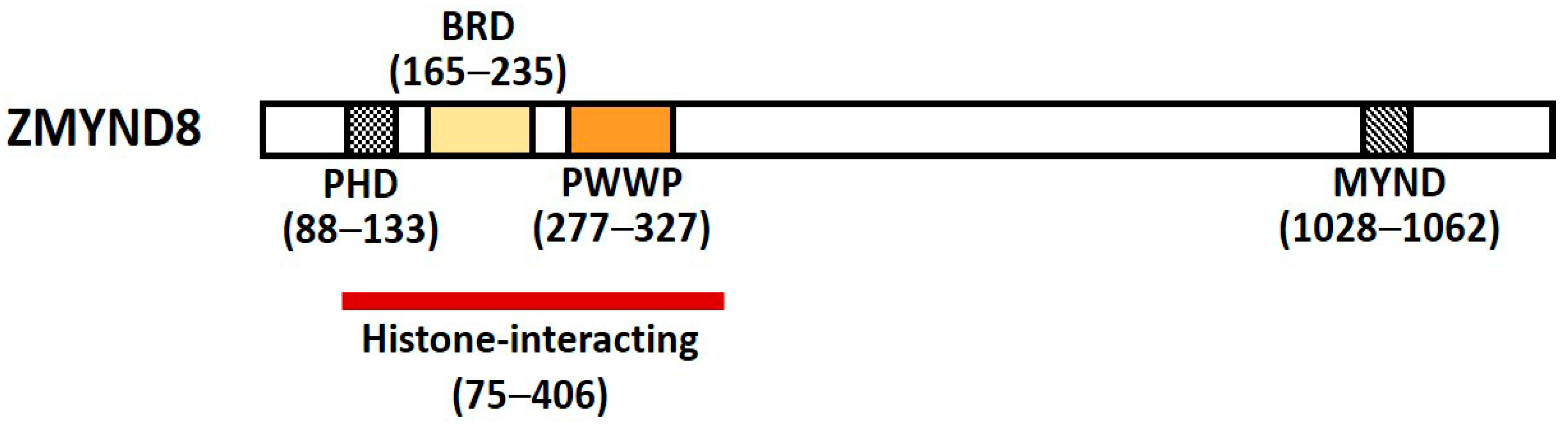
ZMYND8 is a core chromatin reader/effector, with distinct affinity for histone H3 and H4 [2][4][5][6]. The N-terminal PHD–BRD–PWWP domains of ZMYND8 can read several acetyl and methyl lysine residues on histones, including acetyl lysine 14 of H3 (H3K14ac), H4K16ac, and di- and tri-methyl lysine 36 of H3 (H3K36me2, H3K36me3) [6][7]. The PHD–BRD–PWWP domains of ZMYND8 form a stable structural reader ensemble and simultaneously engage histones and DNA, and then ZMYND8 is recruited to the transcriptional sites of the chromatin [7]. Mutation of the reader ensemble may affect the binding of ZMYND8 to histones by disrupting the interaction interface or destabilizing the domain topology [7]. ZMYND8 has been found to regulate gene expression by recognizing dual histone marks [8]. It regulates the expression of all-trans retinoic acid (ATRA)-responsive genes through specific recognition of H3K36me2/H4K16ac [9][10]. It also recognizes H3K4me1/H3K14ac in DU154 and CWR22Rv1 prostate cancer cells [8], as well as H3K36me2/H4K16ac in SH-SY5Y neuroblastoma cells [4][9]. The dual recognition of two different histone modification by ZMYND8 suggests that the two separate conserved domains PWWP and BRD have different affinities towards their cognate histone binding partners [11]. When both H3K36me2 and H4K16ac exist in same histone octamer, the initial binding and recognition of the ZMYND8 to chromatin is considered through H3K36me2 because of its higher association rate, and the stability of the ZMYND8–nucleosome complex relies more on the binding to H4K16ac, due to its lower association rate [11].
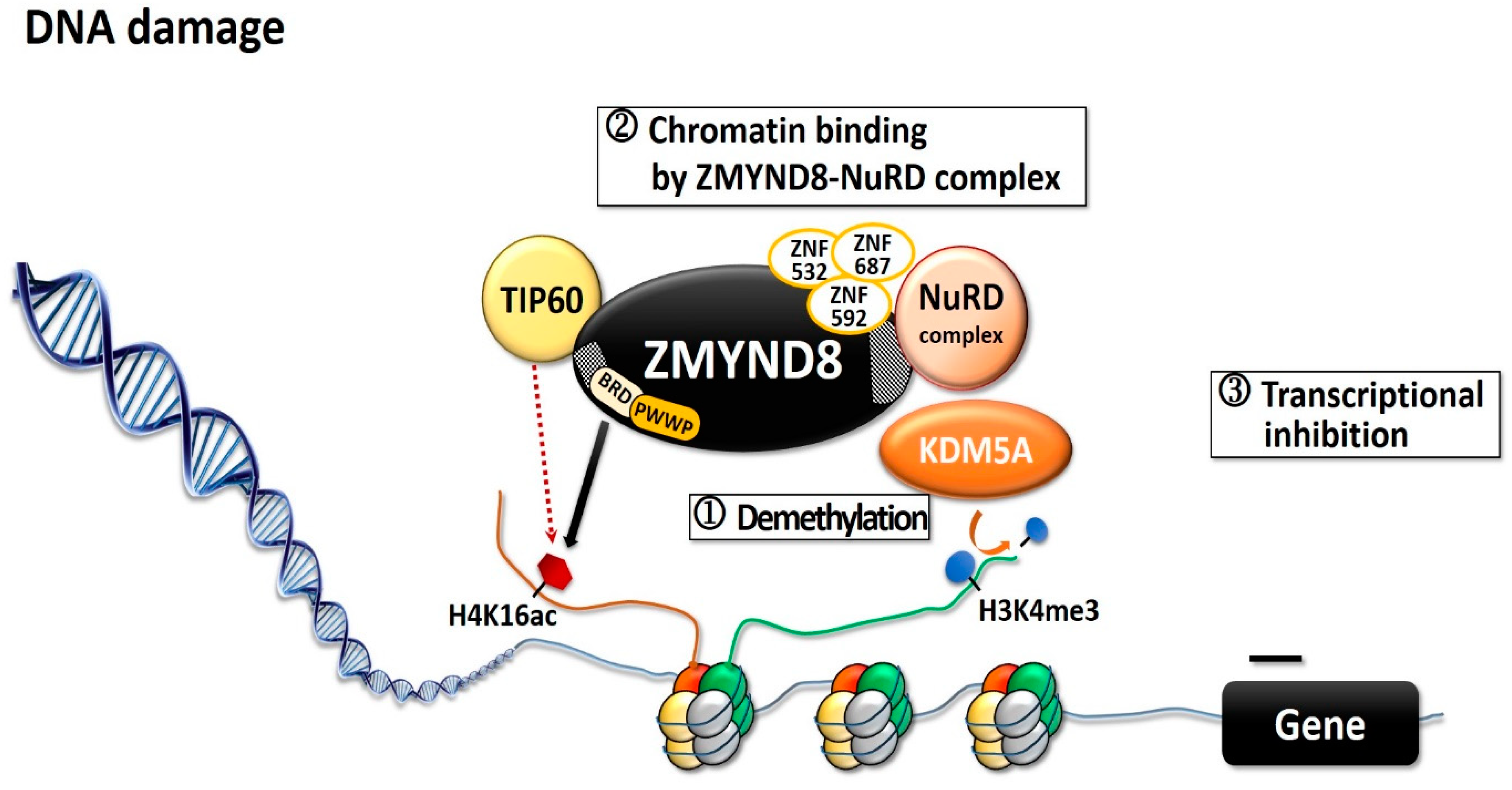
ZMYND8 is involved in transcription activation and in regulating transcription initiation through its interaction with the RNA polymerase II complex [4][12]. Through its putative coiled-coil domain, ZMYND8 forms a homodimer that preferentially associates with positive transcription elongation factor b (P-TEFb) complex, whereas the monomer associates with the chromodomain helicase DNA-binding protein 4 (CHD4) subunit of repressor nucleosome remodeling and deacetylase (NuRD) complex [4][12][13]. ZMYND8 and NuRD share a large number of genome-wide binding sites, mostly in active promoters and enhancers [14]. Both ZMYND8 and CHD4 modulate the expression of many genes and maintain genome integrity; silencing any one of them can alter global gene expression [4][12]. Silencing of ZMYND8 in HeLa cells increases the expression of 331 genes and decreases that of 438 genes [4]. ZMYND8 is important in modulating chromatin integrity and DNA repair [6][15][16]. Upon DNA double-strand break (DSB), histone modifications are altered to accommodate the DNA-damage signaling and the repair [6][15], and BRD2 protein and ZMYND8 are recruited to the DNA damages sites [17]. BRD2 occupies a spatially restricted region extending 2 kb either side of the DSB, and ZMYND8 spreads along the flanking chromatin [17]. The hyperacetylated chromatin domain is required for DBS repair, and the binding of BRD2 to H4ac protects the underlying acetylated chromatin from attack by histone deacetylases, whereas ZMYND8 is a repressor factor which limits transcription during DSB repair [17]. This creates a spatially restricted H4ac/BRD domain which facilitates DSB repair [17]. ZMYND8 interacts with various chromatin-remodeling complexes, histone demethylases/deacetylases, and acetyl transferases, including lysine demethylase 1A (KDM1A), KDM5A, KDM5C, and KDM5D, as well as histone acetyltransferase Tat-interactive protein-60KDa (TIP60) [4][6][8][12][16][18]. KDM5A-dependent demethylation is crucial for the binding of the ZMYND8–NuRD complex to chromatin and its recruitment to the locations of DNA damage (Figure 2) [15]. KDM5A causes H3K4me3 demethylation within chromatin, near the sites of DSB, while ZMYND8, NuRD complex, and KDM5A interact to repress transcription upon DNA damage [6][15]. KDM5A deficiency impairs the transcriptional silencing and the repair of DSBs by homologous recombination [15]. ZMYND8 also interacts with the NuRD complex and TIP60, to mediate DNA repair through homologous recombination [6][16]. In Xenopus embryos, ZMYND8 interacts with RE1-silencing transcription factor corepressor 2 (RCOR2), and together they function as transcriptional repressors in regulating neural differentiation [19]. Both ZMYND8 and p53 play a role in DSB repair [6][15][16]. In several breast cancer cells with distinct p53 genotypes, ZMYND8 loss induced consistent micronucleus formation and DNA-damage response [20]. Additionally, in ZMYND8-depleted human U2OS osteosarcoma cells, laser micro-irradiation induced sustained p53 phosphorylation, which is a DSB marker; in contrast, there was no sustained p53 phosphorylation in control cells [6]. These results imply ZMYND8 and p53 may function independently for repair of DNA damage.
2. Regulation of ZMYND8 to Treat Cancer
Aberrant regulation of gene expression and epigenetic mutations play an important role in carcinogenesis. ZMYND8 is a transcription factor, a histone H3-interacting protein, and a putative chromatin reader/effector, important for regulating transcription in normal cells, and its mutation, altered expression, and fusion with other genes, are associated with development and progression of cancer. Increased expression of ZMYND8 is associated with breast, prostate, colorectal, and cervical cancers, and it is pro-oncogenic in breast and prostate cancers [2][21][22][23]. ZMYND8 is induced during hypoxic conditions, and can promote angiogenesis in zebrafish, as well as in breast and prostate cancers [2][22]. In contrast, lower expression of ZMYND8 is linked to breast, prostate, and nasopharyngeal cancers, and ZMYND8 exerts tumor-suppressive effects on breast and prostate cancers [8][9][24][25]. It suppresses tumor growth by promoting differentiation; inhibiting proliferation, progression of cell cycle, invasiveness, and metastasis; and maintaining the epithelial phenotype in various types of cancer [8][9][24][25]. Although all of these results indicate that ZMYND8 is important in cancer, they are contradictory, and the actual role of ZMYND8 in cancer is still unclear. In addition, the investigations are limited to a few types of cancer, like that of breast and prostate. As ZMYND8 interacts with various transcriptional corepressors, chromatin remodeling complexes, histone demethylase/deacetylase, and acetyl transferase enzymes, the role of ZMYND8 in cancers may be related to not just its direct action, but to multiple factors. Different cell types, fusion with other genes, and the interaction of ZMYND8 with various binding partners may affect the functions of ZMYND8 in cancers. The interaction between ZMYND8 and various binding partners is considered to be crucial in determining whether the function of ZMYND8 is pro-oncogenic or tumor suppressive (Figure 3).
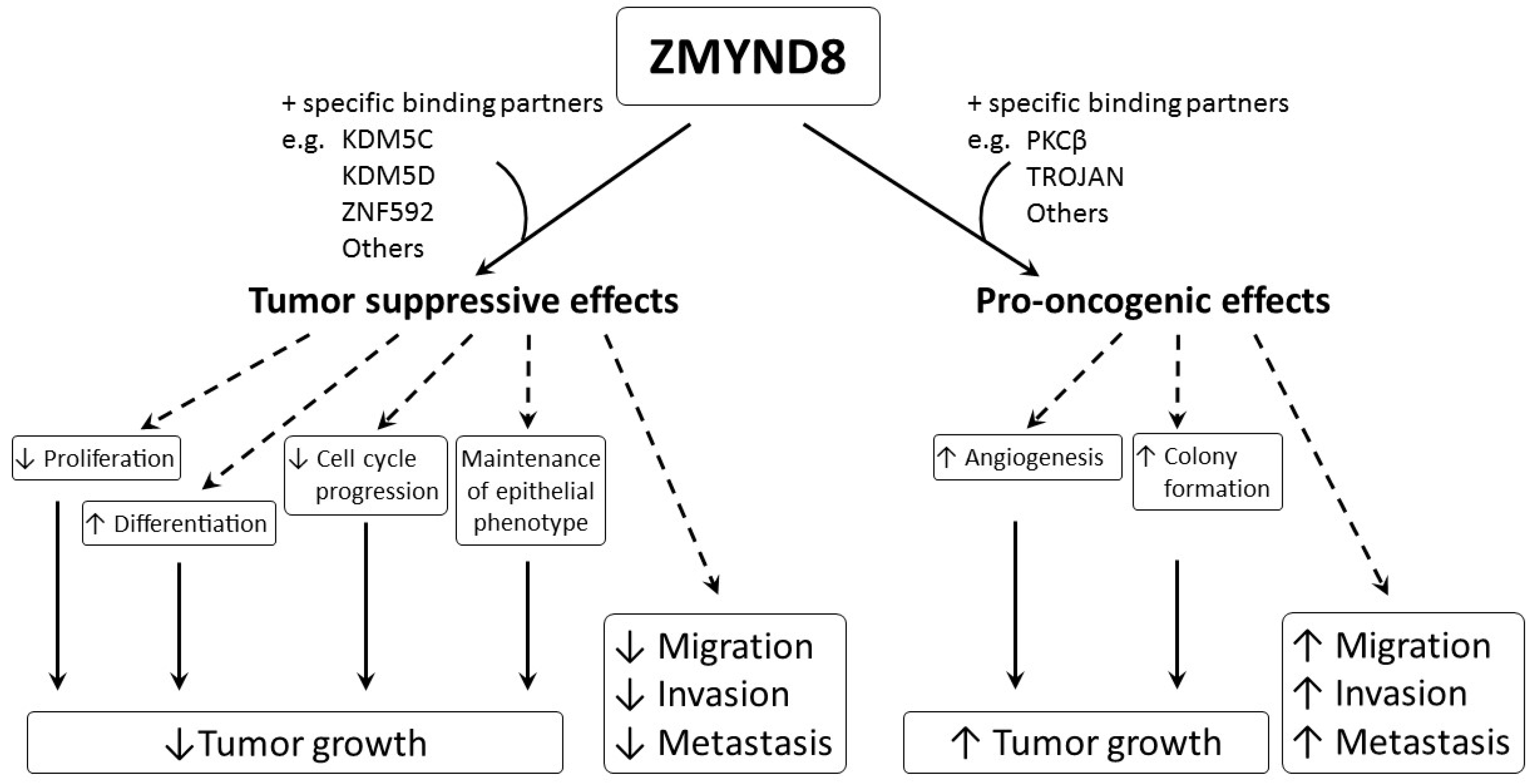
For example, the interaction between ZMYND8 and PKCβ or TROJAN may induce pro-oncogenic effects, while interaction between ZMYND8 and KDM5C, KDM5D, or ZNF592 may cause tumor-suppressive effects. Therefore, more studies are necessary, and they need to be focused not only on ZMYND8, but also on its specific binding partners in different cancer types. Hopefully, in the future, regulation of the expression of ZMYND8 and/or its binding partners may become useful in treating cancer.
This entry is adapted from the peer-reviewed paper 10.3390/molecules26041083
References
- Kuroyanagi, J.; Shimada, Y.; Zhang, B.; Ariyoshi, M.; Umemoto, N.; Nishimura, Y.; Tanaka, T. Zinc finger MYND-type containing 8 promotes tumour angiogenesis via induction of vascular endothelial growth factor-A expression. FEBS Lett. 2014, 588, 3409–3416.
- Adhikary, S.; Sanyal, S.; Basu, M.; Sengupta, I.; Sen, S.; Srivastava, D.K.; Roy, S.; Das, C. Selective recognition of H3.1K36 dimethylation/H4K16 acetylation facilitates the regulation of all-trans-retinoic acid (ATRA)-responsive genes by putative chromatin reader ZMYND8. J. Biol. Chem. 2016, 291, 2664–2681.
- Wen, H.; Li, Y.; Xi, Y.; Jiang, S.; Stratton, S.; Peng, D.; Tanaka, K.; Ren, Y.; Xia, Z.; Wu, J.; et al. ZYMND11 links histone H3.3K36me3 to transcription elongation and tumor suppression. Nature 2014, 508, 263–268.
- Gong, F.; Chiu, L.Y.; Cox, B.; Aymard, F.; Clouaire, T.; Leung, J.W.; Cammarata, M.; Perez, M.; Agarwal, P.; Brodbelt, J.S.; et al. Screen identifies bromodomain protein ZMYND8 in chromatin recognition of transcription-associated DNA damage that promotes homologous recombination. Genes Dev. 2015, 29, 197–211.
- Savitsky, P.; Krojer, T.; Fujisawa, T.; Lambert, J.P.; Picaud, S.; Wang, C.Y.; Shanle, E.K.; Krajewski, K.; Friedrichsen, H.; Kanapin, A.; et al. Multivalent histone and DNA engagement by a PHD/BRD/PWWP triple reader cassette recruits ZMYND8 to K14ac-rich chromatin. Cell Rep. 2016, 17, 2724–2737.
- Li, N.; Li, Y.; Lv, J.; Zheng, X.; Wen, H.; Shen, H.; Zhu, G.; Chen, T.Y.; Dhar, S.S.; Kan, P.Y.; et al. ZMYND8 reads the dual histone mark H3K4me1-H3K14ac to antagonize the expression of metastasis-linked genes. Mol. Cell 2016, 63, 470–484.
- Basu, M.; Khan, M.W.; Chakrabarti, P.; Das, C. Chromatin reader ZMYND8 is a key target of all trans retinoic acid-mediated inhibition of cancer cell proliferation. Biochim. Biophys. Acta 2017, 1860, 450–459.
- Basu, M.; Sengupta, I.; Khan, M.W.; Srivastava, D.K.; Chakrabarti, P.; Roy, S.; Das, C. Dual histone reader ZMYND8 inhibits cancer cell invasion by positively regulating epithelial genes. Biochem. J. 2017, 474, 1919–1934.
- Mukherjee, S.; Sen, S.; Adhikary, S.; Sengupta, A.; Mandal, P.; Dasgupta, D.; Chakrabarti, P.; Roy, S.; Das, C. A novel role of tumor suppressor ZMYND8 in inducing differentiation of breast cancer cells through its dual-histone binding function. J. Biosci. 2020, 45, 2.
- Malovannaya, A.; Lanz, R.B.; Jung, S.Y.; Bulynko, Y.; Le, N.T.; Chan, D.W.; Ding, C.; Shi, Y.; Yucer, N.; Krenciute, G.; et al. Analysis of the human endogenous coregulator complexome. Cell 2011, 145, 787–799.
- Ghosh, K.; Tang, M.; Kumari, N.; Nandy, A.; Basu, S.; Mall, D.P.; Rai, K.; Biswas, D. Positive regulation of transcription by human ZMYND8 through its association with P-TEFb complex. Cell Rep. 2018, 24, 2141–2154.
- Spruijt, C.G.; Luijsterburg, M.S.; Menafra, R.; Lindeboom, R.G.; Jansen, P.W.; Edupuganti, R.R.; Baltissen, M.P.; Wiegant, W.W.; Voelker-Albert, M.C.; Matarese, F.; et al. ZMYND8 Co-localizes with NuRD on Target Genes and Regulates Poly(ADP-Ribose)-Dependent Recruitment of GATAD2A/NuRD to Sites of DNA Damage. Cell Rep. 2016, 17, 783–798.
- Gong, F.; Clouaire, T.; Aguirrebengoa, M.; Legube, G.; Miller, K.M. Histone demethylase KDM5A regulates the ZMYND8-NuRD chromatin remodeler to promote DNA repair. J. Cell Biol. 2017, 216, 1959–1974.
- Gong, F.; Miller, K.M. Double duty: ZMYND8 in the DNA damage response and cancer. Cell Cycle 2018, 17, 414–420.
- Gursoy-Yuzugullu, O.; Chelsea Carman, C.; Price, B.D. Spatially restricted loading of BRD2 at DNA double-strand breaks protects H4 acetylation domains and promotes DNA repair. Sci. Rep. 2017, 7, 12921.
- Eberl, H.C.; Spruijt, C.G.; Kelstrup, C.D.; Vermeulen, M.; Mann, M. A map of general and specialized chromatin readers in mouse tissues generated by label-free interaction proteomics. Mol. Cell 2013, 49, 368–378.
- Zeng, W.; Kong, Q.; Li, C.; Mao, B. Xenopus RCOR2 (REST corepressor 2) interacts with ZMYND8, which is involved in neural differentiation. Biochem. Biophys. Res. Commun. 2010, 394, 1024–1029.
- Wang, Y.; Luo, M.; Chen, Y.; Wang, Y.; Zhang, B.; Ren, Z.; Bao, L.; Wang, Y.; Wang, J.E.; Fu, Y.X.; et al. ZMYND8 expression in breast cancer cells blocks T-lymphocyte surveillance to promote tumor growth. Cancer Res. 2020, 81, 174–186.
- Bierkens, M.; Krijgsman, O.; Wilting, S.M.; Bosch, L.; Jaspers, A.; Meijer, G.A.; Meijer, C.J.; Snijders, P.J.; Ylstra, B.; Steenbergen, R.D. Focal aberrations indicate EYA2 and hsa-miR-375 as oncogene and tumor suppressor in cervical carcinogenesis. Genes Chromosomes Cancer 2013, 52, 56–68.
- Chen, Y.; Zhang, B.; Bao, L.; Jin, L.; Yang, M.; Peng, Y.; Kumar, A.; Wang, J.E.; Wang, C.; Zou, X.; et al. ZMYND8 acetylation mediates HIF-dependent breast cancer progression and metastasis. J. Clin. Invest. 2018, 128, 1937–1955.
- Chen, J.; He, Q.; Wu, P.; Fu, J.; Xiao, Y.; Chen, K.; Xie, D.; Zhang, X. ZMYND8 expression combined with pN and pM classification as a novel prognostic prediction model for colorectal cancer: Based on TCGA and GEO database analysis. Cancer Biomark. 2020, 28, 201–211.
- Jin, X.; Xu, X.E.; Jiang, Y.Z.; Liu, Y.R.; Sun, W.; Guo, Y.J.; Ren, Y.X.; Zuo, W.J.; Hu, X.; Huang, S.L.; et al. The endogenous retrovirus-derived long noncoding RNA TROJAN promotes triple-negative breast cancer progression via ZMYND8 degradation. Sci. Adv. 2019, 5, eaat9820.
- Shen, H.; Xu, W.; Guo, R.; Rong, B.; Gu, L.; Wang, Z.; He, C.; Zheng, L.; Hu, X.; Hu, Z.; et al. Suppression of enhancer overactivation by a RACK7-histone demethylase complex. Cell 2016, 165, 331–342.
