There are millions of reported cases of foodborne illness in developed and developing countries annually making identification of foodborne pathogens a global concern. Testing for foodborne pathogens is essential to ensuring a safe food supply, quality assurance in the food industry, treating illness, and controlling outbreaks. In this article, several methods for the detection and identification of foodborne pathogens are discussed and a triplex PCR high-resolution melt assay for detecting Vibrio parahaemolyticus, Shigella flexneri, and Escherichia coli is presented.
- pathogen
- immunoassay
- ELISA
- biosensor
- PCR
- DNA sequencing
- Vibrio
- Shigella
- E. coli
There are millions of reported cases of foodborne illness in developed and developing countries annually making identification of foodborne pathogens a global concern. The United States Centers for Disease Control and Prevention (CDC) reports that there are 48 million cases of foodborne diseases, affecting approximately 1 in 6 people annually in the United States; these result in 128,000 hospitalizations and 3000 deaths[1]. Contaminated water can harbor pathogens or their toxins[1]. Uncooked and ready-to-eat foods including meats, cheeses, eggs, salad greens, vegetables, fruits, nuts, and raw seafood and shellfish are most likely to cause infection[1]. Flour, cereals, soy nut butter, tofu, tahini and pasta salad have also been affected[1]. Testing is essential to ensuring a safe food supply, performing quality assurance in the food industry, treating illness, and controlling outbreaks. Several methods for the detection and identification of foodborne pathogens have been developed and published and are in use by hospitals, clinical laboratories and health departments.
More than 250 foodborne pathogens have been identified and include bacterial and viral pathogens, parasites and worms[2]. Bacterial pathogens include Salmonella, Listeria, Escherichia, Campylobacter, Vibrio, Bacillus, Shigella, Staphylococcus, Clostridium, and Yersinia[1][2]. Viral pathogens include Norovirus, Rotavirus, and Hepatitis A[1][2]. Other pathogens include Giardia, Cyclospora, Crysptosporidium, and Toxoplasmosis parasites and Trichinella worms[1][2]. Additionally, several species including Shiga toxin-producing E. coli (STEC), botulinum toxin produced by Clostridium botulinum among other strains, produce toxins that can be ingested via food and water[1][2]. Campylobacter is the most commonly identified agent in foodborne infections since 2013 in the United States[1]. Salmonella infections are the second most common type of foodborne pathogen infection; the most common serotypes are Enteritidis, Newport and Typhimurium[1]. Shigella and Cryptosporidium are the third and fourth most common pathogens, respectively, in the U.S.[1]. Due to the rise of Vibrio in coastal waters, cases involving this pathogen are on the rise[1]. Because of new testing methods, more cases of Vibrio and Yersinia are now being diagnosed including V. parahaemolyticus, V. vulnificus, and Y. enterocolitica.[1] E. coli O157:H7, E. coli O13:H25, B. cereus, S. aureus also cause a large number or infections[1]. Foodborne illness cases caused by C. botulinum, Listeria sp., E. coli, and Vibrio sp. are more likely to lead to hospitalization, according to the CDC[1].
In order to properly treat infections and prevent and control outbreaks, the causative agent needs to be rapidly detected and identified. In the case of an outbreak identification methods are essential to identifying the source and index case. For routine screening, methods are needed to identify the possible presence of a pathogen in foods and water. Rapid methods are essential to reduce time in an emergency response and keep labor costs manageable[3].
Several methods of detecting and identifying foodborne pathogens have been developed and tested[3][4]. The oldest method is culturing bacteria on agar plates, isolating the pathogen, and staining[3][4]. The shape, color and other morphological characteristics are examined under a microscope to yield an identification or tentative identification. Although this an inexpensive and reliable method and remains the gold standard, it is slow, labor-intensive, and may lead to inconclusive results as not all bacteria can be cultured. In addition, some bacteria are difficult to culture or will only grow under specific conditions and the culture method cannot detect the production of bacterial toxins. Culturing cannot distinguish phenotypically similar species[5].
Fluorescein isothiocyanate can be used to detect enzyme activity in viable but non-culturable (VBNC) cells[4]. Even if bacteria are culturable, growth may take three to seven days[3][4]. Stains can be used to screen and differentiate bacteria, although several may need to be used to make a tentative identification. For example, the Gram stain stains bacteria with teichoic acids in the cell wall purple[3]. However, there are several Gram positive bacteria that result in a pink color[3]. Further, even with staining, not all bacteria can be identified microscopically. Differentiating strains may require additional testing. Newer methods for detection and differentiation include immunological, biosensor-based, nucleic-acid based assays, and mass spectrometry proteomic and metabolomics assays[3][4][5][6][7][8][9].
Immunological assays include latex agglutination testing, enzyme-linked immunosorbent assays (ELISA), lateral flow immunochromatographic testing, and immunofluorescence assays. Immunological tests rely on antibody-antigen reaction testing[4][6][8]. Tests may employ polyclonal or monoclonal antibodies; the specificity and sensitivity of these tests depends upon the binding affinity of the antigen to the antibody. While monoclonal antibodies yield the best specificity, tests employing these are also the most expensive. Immunological tests can be very sensitive, rapid and inexpensive depending upon the quantity of antibody used. Latex agglutination tests are rapid tests that utilize latex beads that are coated with a pathogen-specific antigen or antibody. Lateral flow immunochromatographic tests are rapid, inexpensive and suitable for on-site detection. These include dipstick and immunochromatographic strip assays. However, many of these tests are not selective for a specific pathogen and will yield a positive result with many species. ELISA assays may employ color-generating reporters and immobilized capture antibodies that indicate the presence of a pathogen due to the binding of one of its specific antigens[4][6][8].
Biosensor-based methods include electrochemical, magnetic, mass-based, micromechanical, optical and thermometric methods that convert biological interactions into a measurable signal[6][8]. One optical biosensor employs surface plasmon resonance (SPR) using reflectance spectroscopy[6][8]. Pathogen binding to a metal surface containing an immobilized receptor is detected using the resulting refractive index. Electrochemical biosensors can detect pathogens due to the changes in current, voltage or conductance[6][8].
Nucleic acid-based assays include end-point polymerase chain reaction (PCR) and gel analysis, real-time PCR with a fluorescence-based assay, real-time PCR with high resolution melt (HRM), loop-mediated isothermal amplification (LAMP) assays, oligonucleotide DNA microarray assays, 16S rRNA sequencing, and whole genome sequencing[5][6][9]. Nucleic acid-based methods can be used to detect specific DNA and RNA sequences including both bacterial and toxin-specific genes by employing highly specific oligonucleotide probes or primers. Nucleic acid-based methods have several advantages over other methods. They can be rapid, sensitive, inexpensive, suitable for on-site detection, automated, selective, and high-throughput. One drawback is that nucleic acid-based tests detect live and dead cells, the latter of which will not cause infection. And while they can detect toxin-encoding genes, they cannot detect toxins. However, PCR-based assays are very sensitive and can detect a single bacterial or viral pathogen. Whole genome sequencing can lead to unequivocal determination of the pathogen when other methods result in unanswered questions. Nucleic-acid based methods are flexible and can be used to analyze a range of pathogens. In addition to sequencing and genomics methods, omics methods including transcriptomics, proteomics, lipidomics, and metabolomics are also being used to detect foodborne pathogens[10].
Multiple genes can be detected with a single assay when the primers are combined into a multiplex. Multiplex assays have been developed using fluorescent dye-labeled primers, Taq Man and other reporter-quencher fluorescence assays, and fluorescent DNA intercalating dyes such as LC Green PLUS[4][5]. Multiplex PCR assays followed by high-resolution melt analysis have been demonstrated to detect and identify pathogens through the use of specific primers that target and amplify each pathogen and enable their identification by different melt temperatures[5]. As an example, Figure 1 shows the melt of a PCR HRM multiplex assay for E. coli, S. flexneri and V. parahaemolyticus. Each bacteria can be detected and identified simultaneously by their unique curves, as shown, and quantified by the threshold cycle. The multiplex reaction was comprised of 8 µL 2.5X LightScanner® master mix (BioFire Defense, Salt Lake City, UT), 1 µL each of 1 ng/µL E. coli (ATCC® 700926™), V. parahaemolyticus (ATCC® 17802-D5™), and S. flexneri (ATCC® 29903™) DNA, 2 µL of 5 µM E. coli forward primer (5’- TCCTGGATTGAGGTGCTTTATC-3’), 1 µL of 5 µM E. coli reverse primer (5’- CTACGGAGACCTGGGTAATTCC-3’), 1 µL of 5 µM V. parahaemolyticus forward primer (5’-ACTGGATTTCGCTTTGCCCTCAATGAA-3’), 1 µL of 5 µM V. parahaemolyticus reverse primer (5’-GTTCTGAGTTCGATAACCTCTTGTGTGGATTAAG-3’), 0.5 µL of 5 µM S. flexneri forward primer (CGAAACTCTAGCCGAAAAACATAGCG-3’), 0.5 µL of 5 µM S. flexneri reverse primer (5’- GTGTCTTTAACGTGGAAGTAATCATCG-3’), and 3 µL of PCR-grade water. The DNA was amplified using the following temperature program using a Rotor-Gene Q real-time PCR instrument (Qiagen, Germantown, MD): 95ºC hold for 2 minutes; cycling 40 times at 95ºC for 20 seconds, 62ºC for 20 seconds with a touchdown of 2 cycles of 0.5ºC, and 72ºC for 20 seconds; and hold at 45ºC for 1 minute. Melt analysis was performed by heating from 65ºC to 95ºC, rising by 0.2 degrees each step, waiting for 90 seconds of pre-melt conditioning on the first step, and waiting for 2 seconds for each step afterwards, with gain optimized. Assays for these and other pathogens continue to be an area of ongoing research and development.
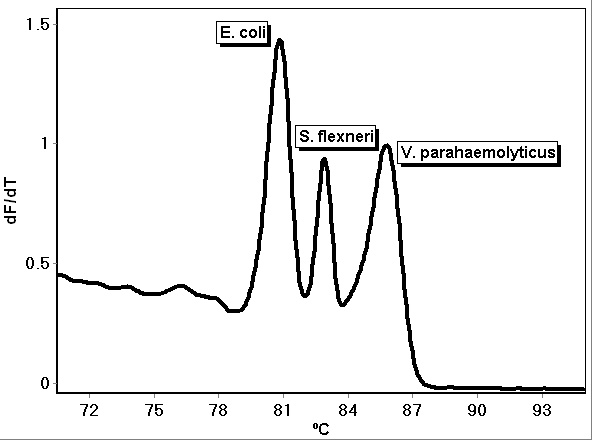
Figure 1. PCR multiplex assay melt curves for E. coli, S. flexneri and V. parahaemolyticus.
References
- Estimates of Foodborne Illness in the United States . Centers for Disease Control and Prevention. Retrieved 2019-11-11
- Timothy L Switaj; Kelly J Winter; Scott R Christensen; Diagnosis and Management of Foodborne Illness. American family physician 2015, 92, 358-365, https://www.aafp.org/afp/2015/0901/p358.html.
- Kelly M. Elkins . Introduction to Forensic Chemistry; Taylor and Francis / CRC Press : Boca Raton, FL, 2018; pp. 251-286.
- B. Priyanka; Rajashekhar K. Patil; Sulatha Dwarakanath; A review on detection methods used for foodborne pathogens. Indian Journal of Medical Research 2015, 144, 327-338, 10.4103/0971-5916.198677.
- Kelly M. Elkins; Anjelica C.U. Perez; Katherine C. Sweetin; Rapid and inexpensive species differentiation using a multiplex real-time polymerase chain reaction high-resolution melt assay. Analytical Biochemistry 2016, 500, 15-17, 10.1016/j.ab.2016.01.013.
- Jodi Woan-Fei Law; Nurul-Syakima Ab Mutalib; Kok Gan Chan; Learn-Han Lee; Rapid methods for the detection of foodborne bacterial pathogens: principles, applications, advantages and limitations. Frontiers in Microbiology 2015, 5, 770, 10.3389/fmicb.2014.00770.
- Snehal R. Jadhav; Rohan M. Shah; Avinash V. Karpe; Paul D. Morrison; Konstantinos Kouremenos; David J. Beale; Enzo A. Palombo; Detection of Foodborne Pathogens Using Proteomics and Metabolomics-Based Approaches.. Frontiers in Microbiology 2018, 9, 3132, 10.3389/fmicb.2018.03132.
- Md Eshrat E. Alahi; Subhas Chandra Mukhopadhyay; Detection Methodologies for Pathogen and Toxins: A Review. Sensors 2017, 17, 1885, 10.3390/s17081885.
- Piumie Rajapaksha; Aaron James Elbourne; Sheeana Gangadoo; Robert Brown; Daniel Cozzolino; James Chapman; A review of methods for the detection of pathogenic microorganisms. The Analyst 2018, 144, 396-411, 10.1039/c8an01488d.
- David I. Ellis; Howbeer Muhamadali; Malama Chisanga; Royston Goodacre; Omics Methods For the Detection of Foodborne Pathogens. Encyclopedia of Food Chemistry 2018, 1, 364-370, 10.1016/b978-0-08-100596-5.21793-9.
