With the development of next generation sequencing technologies in recent years, it has been demonstrated that many human infectious processes, including chronic wounds, cystic fibrosis, and otitis media, are associated with a polymicrobial burden. Research has also demonstrated that polymicrobial infections tend to be associated with treatment failure and worse patient prognoses. Despite the importance of the polymicrobial nature of many infection states, the current clinical standard for determining antimicrobial susceptibility in the clinical laboratory is exclusively performed on unimicrobial suspensions. There is a growing body of research demonstrating that microorganisms in a polymicrobial environment can synergize their activities associated with a variety of outcomes, including changes to their antimicrobial susceptibility through both resistance and tolerance mechanisms.
- polymicrobial
- antimicrobial susceptibility
- synergism
- interactions
- clinical
- diagnostics
1. Introduction
With recent developments in sequencing technologies, it has been shown that many infections can be polymicrobial in nature, potentially leading to worse patient outcomes. Current routine clinical models, however, focus on unimicrobial culture-based methods to determine the causative agent. It is now well-known that many chronic infections are often polymicrobial in nature [1]. Often, chronic infections are difficult to treat as polymicrobial interactions can lead to decreased antibiotic efficacy [1]. Recently developed next generation sequencing (NGS) technologies have led to increased understanding of polymicrobial infections, as they have helped to eliminate culture-bias in microbial identification. Culture-based microbial identification is a common method used in clinical laboratories to identify the microorganisms in samples [2]. However, it has been shown that certain species of microbes are often lost in culture, either due to competition from other microorganisms, the need for specific growth requirements, or other causes. NGS has proven effective to accurately recount the composition of microbial communities that would normally take multiple culture attempts using different differential and/or selective media and non-routine culture methods [3]. With NGS helping to more accurately identify the microorganisms in samples, it is now being demonstrated that more infections are polymicrobial than was previously recognized. Studies examining the polymicrobial nature of infectious processes are discussed below, and unless otherwise noted, the cited studies determined the polymicrobial consortia utilizing NGS methods.
2. Mechanisms of Polymicrobial Synergism
There are many complex mechanisms pathogenic microbes can use to not simply survive, but also thrive in polymicrobial infections. Not only can pathogens work with each other, but they can work with commensal organisms, and even the host [4] (Figure 2). Many polymicrobial infections contain microorganisms that work cooperatively.
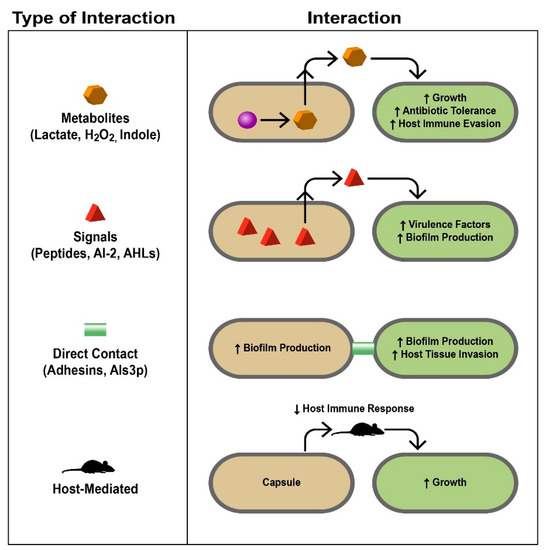
Figure 2. Mechanistic bases of polymicrobial interactions. Different interactions discussed in this review are summarized here. The left column lists the types of interactions and how they are mediated with specific examples, some of which are discussed in the text. The right column demonstrates how the interaction occurs and the response of the different microbes involved and the infected host. Signals, proteins, metabolites, and even the host immune system serve as liaisons between different microbes, allowing complex interactions to occur that impact the environments in which they live. During infection, these interactions ultimately lead to polymicrobial synergy and are therefore detrimental to the host (AI-2, autoinducer-2; AHLs, acyl-homoserine lactones). Taken from [4], reprinted with permission from Journal of Microbiology (Springer Nature).
2.1. Metabolites
In addition to biofilm formation, in which a hierarchical, often polymicrobial bacterial community can be established according to their nutrient and oxygen needs, metabolite cross-feeding, in which one species utilizes the metabolic pathway end-products of another species, allows for increased bacterial survival and growth This phenomenon has been extensively researched in oral bacteria, especially dental plaque [5]. An example of a cooperative relationship between bacteria is the mixed biofilm formed by Veillonella atypica and S. gordonii; S. gordonii produces lactate, which is then consumed by V. atypica; S. gordonii in turn responds to a diffusible molecule produced by V. atypica to produce amylase [5]. Ramsey et al. demonstrated that Aggregatibacter actinomycetemcomitans pathogenesis is enhanced when co-cultured with S. gordonii; S. gordonii produces L-lactate as its primary metabolite, which is then utilized by A. actinomycetemcomitans as a carbon source necessary for vigorous growth [6]. The fungus Candida albicans has been shown to possess a polymicrobial relationship with multiple bacterial species. It helps to create an anoxic environment allowing for gingival inflammation by the anaerobic bacteria Porphyromonas gingivalis [7]. Regarding metabolite cross-feeding, C. albicans metabolizes glucose produced by the bacteria Streptococcus mutans when it breaks down sucrose;this metabolization of glucose creates an acidic environment in which both the bacterial and fungal species thrive [7].
2.2. Signals
Quorum sensing, a mechanism of cell–cell communication based on population density mediated by signaling molecules, is another method of which microbes can benefit from polymicrobial infections. Microbial “cheaters” can use signals created by other microorganisms in order to avoid having to produce signals of their own. By listening in to the signals produced by other microbes (termed “eavesdropping”), these cheaters can perform tasks and react to environmental changes without having to expend energy on producing their own signals [8].
2.3. Direct Contact
Direct contact can contribute to polymicrobial synergy as microorganisms directly interact with each other. An example of this phenomenon is S. epidermidis protecting the fungus C. albicans from the antifungal fluconazole by producing an extracellular slime, increasing the content of their mixed biofilm and preventing contact of the antifungal drug with the fungus [9].
2.4. Host-Mediated
Host-mediated mechanisms of synergy are commonly seen in polymicrobial infections involving a virus, but can also be seen in polymicrobial infections involving bacteria and fungi. Host-mediated methods of synergism include mechanisms such as immune system modulation, in which a virus can decrease the host’s immune response, allowing for increased proliferation of other microbes. Often, other species in polymicrobial infections can benefit from this phenomenon, allowing for increased growth and more antibiotic resistance as the bacteria can focus on surviving the antibiotic instead of fighting the host’s immune system [10].
3. Polymicrobial Synergism and Its Impact on Antimicrobial Susceptibility
The literature surrounding the effect of polymicrobial synergism on antimicrobial susceptibility is, however, far from comprehensive. There are three main categories of these interactions investigated in this review—bacteria–bacteria, bacteria–fungus, and bacteria–viral, and each is less well elucidated than the last. The vast majority of the available literature is limited by two major factors—the limited number of species used in the experiments and the frequent use of non-standard conditions and models. Of all the polymicrobial combinations, the perhaps most commonly investigated relationship is the synergistic interaction of P. aeruginosa and S. aureus, two common colonizers of both the chronic wound and cystic fibrosis microbiomes. Polymicrobial interactions between bacteria–bacteria and bacteria–fungi with evidence of changes to antimicrobial efficacy are summarized in Table 1.
Table 1. Summarized selection of changes to antimicrobial efficacy in polymicrobial conditions. This figure summarizes the observed effects of a polymicrobial condition on antibiotic efficacy in both bacteria–bacteria and bacteria–fungus interactions across a variety of species and antimicrobial combinations.
| Citation | Organisms Studied | Observation |
|---|---|---|
| Bacteria–Bacteria | ||
| Hoffman et al. [11] | P. aeruginosa, S. aureus | 2x increase in tolerance to tobramycin |
| Orazi and O’Toole [12] | P. aeruginosa, S. aureus | Increased tolerance to B-lactam, glycopeptide, aminoglycoside, macrolide, tetracycline classes |
| Lebrun et al. [13] | P. aeruginosa, S. aureus | Increased tolerance to rifamycin, vancomycin, penicillin, cycloserine |
| DeLeon et al. [14] | P. aeruginosa, S. aureus | Increased tolerance to gentamicin, tetracycline |
| Vega et al. [15] | E. coli, S. typhimurium | Increased tolerance to ciprofloxacin |
| Adamowicz et al. [16] | E. coli, S. typhimurium | Increased tolerance to tetracycline |
| Tavernier et al. [17] | P. aeruginosa, S. aureus, S. typhimurium | Increased tolerance to vancomycin |
| Bacteria–Fungus | ||
| Harriot et al. [18] | C. albicans, S. aureus | Increase in biofilm formation |
| Todd et al. [7] | C. albicans, S. aureus | Lethality increase |
| Kean et al. [19] | C. albicans, S. aureus | 4x increased tolerance to miconazole |
| Kong et al. [20] | C. albicans, S. mutans | Increased tolerance to fluconazole, response to farnesol |
| Förster et al. [9] | C. albicans, S. epidermidis | Increased tolerance to fluconazole, “slime” production |
3.1. Bacteria–Bacteria Interactions
Across a variety of experimental conditions and antibiotic classes, the co-culturing of P. aeruginosa (PA) and S. aureus (SA) has been shown to result in reduced antimicrobial susceptibility for S. aureus. Hoffman et al. [11], for example, demonstrated a doubling of the tobramycin MIC in SA when exposed to the PA exoproduct 4-hydroxy-2-heptylquinoline-N-oxide (HQNO). Orazi and O’Toole [12] exposed SA to the supernatant of a PA culture, and after testing some 240 antibiotics, found that “cell wall synthesis inhibitors and protein synthesis inhibitors…includ[ing] multiple representatives from the B-lactam, glycopeptide, aminoglycoside, macrolide, and tetracycline classes” showed decreased efficacy. Even as far back as 1978, Lebrun et al. [13] demonstrated a similar effect in a simple co-culture of the two species, with decreased susceptibility to rifamycin, vancomycin, penicillin, and cycloserine. While other studies had limited the direct exposure of PA and SA due to their competition in vitro, DeLeon et al. [14] used a wound-like media to allow for their co-culturing, which is consistent with the clinically observed conditions of several infectious processes, and also showed significant increases in tolerance for SA to both gentamicin and tetracycline. While this increase in tolerance to a wide variety of antibiotics has been more fully explored in this pair than that of other species, this is not, however, the only bacteria–bacteria interaction that has been studied. Vega et al. [15] demonstrated an increase in the tolerance of S. typhimurium to ciprofloxacin when exposed to the E. coli signaling molecules indole and tryptophan, and in a metabolically cross-feeding community, E. coli saw a doubling of MIC when exposed to tetracycline, even as the most susceptible organism in the culture [16]. Increases in tolerance to vancomycin were also observed in polymicrobial biofilm models using Staphylococcus anginosus, which were attributed to cell wall thickening, a mechanism which has been demonstrated elsewhere as an effect of synergistic interactions among bacteria [17]. Interactions between PA and anaerobes isolated from CF patient sputum have also been demonstrated to increase PA’s tolerance to piperacillin [21]. Though the largest body of research demonstrating changes in antimicrobial tolerance exists in this field, these interactions are certainly not limited to a single kingdom.
3.2. Bacteria–Fungus Interactions
Bacteria–fungus interactions, and their effect on polymicrobial susceptibility, are far less well examined than that of bacteria–bacteria interactions, but a body of research does exist. Of these, the relationship between Candida albicans and S. aureus has been examined in some detail, and along with increased biofilm formation [18] and an increase in synergistic lethality [7], an increased tolerance to both antibiotics and antifungal compounds has been observed when the organisms are in co-culture. Kean et al. [19] demonstrated a fourfold increase in tolerance to miconazole for S. aureus when cultured with C. albicans, an effect which has been supported by a similar decrease in C. albicans sensitivity to fluconazole when cultured with S. mutans. Examinations of the relationship between these organisms attributed the cause of the increased tolerance to a higher expression of drug efflux pumps on the part of S. aureus. In addition, the researchers demonstrated the ability of that same organism to respond to the quorum sensing molecule farnesol produced by C. albicans [20]. S. epidermidis has also been shown to have a protective effect on C. albicans by the production of an extracellular “slime”, which decreases the efficacy of fluconazole on the fungus [9]. Given that bacterial–fungal co-infection can occur across a wide number of body systems, including the enteric, respiratory, and oral cavity, this relative paucity of studies determining what changes occur in the respective organisms’ MICs demonstrates a gap in the available literature.
3.3. Bacteria–Virus Interactions:
It is in this last category of interactions that the research is the most sparse, when focusing specifically on changes to antibiotic susceptibility. Though a variety of interactions between a number of respiratory viruses and bacteria [22][23], HIV and Mycobacterium tuberculosis [24], or the Epstein-Barr virus and Porphyromonas endodontalis or P. gingivalis have been described in detail [25], they have focused largely on the interactions occurring through the indirect mediation of host immune system activity, rather than a direct bacteria–virus interaction resulting in a change in MIC in constituent infectious bacteria. This trend is common in the available literature and viral–bacterial interaction groups. For instance, in a study of otitis media among pediatric patients, with one group having only a bacterial infection but the other having both bacterial and viral infections of the middle ear, differences in total clinical treatment success or failure were not statistically significant. However, a strong increase in the failure of initial antibiotic treatment was observed among the combined viral–bacterial infections, with 50% of the co-infected (bacterial and viral) group displaying failure compared to only 13% of the bacteria-only group [10]. In addition, of those in the bacteria-only failure group, 75% had bacteria with pre-existing resistance to the prescribed antibiotic, whereas 66% of the co-infected group’s bacteria were susceptible when evaluated using the Kirby-Bauer disk diffusion method [10]. This would seem to indicate that bacteria–virus co-infection is producing an effect on the antimicrobial susceptibility of the bacteria, but it is far less certain whether or not that effect relates more to the possible indirect immune suppression of the host due to the concomitant viral infection or due to the direct interactions of those two microorganisms with each other. It has been demonstrated that the presence of an Influenza A infection significantly decreases the physical penetration of antibiotics in a chinchilla model of otitis media, which may explain the number of failures of antibiotic treatment in that infectious process [26]. Outside of clinical studies, viruses have been shown to be able to increase the biofilm formation of P. aeruginosa, with a concomitant attendant effect on antimicrobial susceptibility [27], but it is less clear what effect the interaction of bacteria and viruses has, in a planktonic form, on antimicrobial susceptibility. In addition, the relevant literature has described direct bacteria–virus interactions within the context of enteric and respiratory infections, leading to increased bacterial and viral adherence and stability, which could suggest changes to bacterial MIC, although that has yet to be definitively determined [28][29][30][31]. Given the multiplicity of infections where viruses may be members of a polymicrobial community [27], this phenomenon certainly deserves more research.
This entry is adapted from the peer-reviewed paper 10.3390/pathogens10020144
References
- Orazi, G.; O’Toole, G.A. “It Takes a Village”: Mechanisms Underlying Antimicrobial Recalcitrance of Polymicrobial Biofilms. J. Bacteriol. 2019, 202.
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 11th ed.; CLSI: Wayne, PA, USA, 2018.
- Cummings, L.A.; Hoogestraat, D.R.; Rassoulian-Barrett, S.L.; Rosenthal, C.A.; Salipante, S.J.; Cookson, B.T.; Hoffman, N.G. Comprehensive evaluation of complex polymicrobial specimens using next generation sequencing and standard microbiological culture. Sci. Rep. 2020, 10, 5446.
- Murray, J.L.; Connell, J.L.; Stacy, A.; Turner, K.H.; Whiteley, M. Mechanisms of synergy in polymicrobial infections. J. Microbiol. 2014, 52, 188–199.
- Short, F.L.; Murdoch, S.L.; Ryan, R.P. Polybacterial human disease: The ills of social networking. Trends Microbiol. 2014, 22, 508–516.
- Ramsey, M.M.; Rumbaugh, K.P.; Whiteley, M. Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLoS Pathog. 2011, 7, e1002012.
- Todd, O.A.; Peters, B.M. Candida albicans and Staphylococcus aureus Pathogenicity and Polymicrobial Interactions: Lessons beyond Koch’s Postulates. J. Fungi 2019, 5, 81.
- Dixon, E.F.; Hall, R.A. Noisy neighbourhoods: Quorum sensing in fungal-polymicrobial infections. Cell. Microbiol. 2015, 17, 1431–1441.
- Förster, T.M.; Mogavero, S.; Dräger, A.; Graf, K.; Polke, M.; Jacobsen, I.D.; Hube, B. Enemies and brothers in arms: Candida albicans and gram-positive bacteria. Cell. Microbiol. 2016, 18, 1709–1715.
- Chonmaitree, T.; Owen, M.J.; Howie, V.M. Respiratory viruses interfere with bacteriologic response to antibiotic in children with acute otitis media. J. Infect. Dis. 1990, 162, 546–549.
- Hoffman, L.R.; Déziel, E.; D’Argenio, D.A.; Lépine, F.; Emerson, J.; McNamara, S.; Gibson, R.L.; Ramsey, B.W.; Miller, S.I. Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2006, 103, 19890–19895.
- Orazi, G.; O’Toole, G.A. Pseudomonas aeruginosa Alters Staphylococcus aureus Sensitivity to Vancomycin in a Biofilm Model of Cystic Fibrosis Infection. mBio 2017, 8.
- Lebrun, M.; de Repentigny, J.; Mathieu, L.G. Diminution of the antibacterial activity of antibiotics in cultures and in experimental mixed infections. Can. J. Microbiol. 1978, 24, 154–161.
- DeLeon, S.; Clinton, A.; Fowler, H.; Everett, J.; Horswill, A.R.; Rumbaugh, K.P. Synergistic Interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an In Vitro Wound Model. Infect. Immun. 2014, 82, 4718–4728.
- Vega, N.M.; Allison, K.R.; Samuels, A.N.; Klempner, M.S.; Collins, J.J. Salmonella typhimurium intercepts Escherichia coli signaling to enhance antibiotic tolerance. Proc. Natl. Acad. Sci. USA 2013, 110, 14420–14425.
- Adamowicz, E.M.; Flynn, J.; Hunter, R.C.; Harcombe, W.R. Cross-feeding modulates antibiotic tolerance in bacterial communities. ISME J. 2018, 12, 2723–2735.
- Tavernier, S.; Sass, A.; De Bruyne, M.; Baeke, F.; De Rycke, R.; Crabbé, A.; Vandecandelaere, I.; Van Nieuwerburgh, F.; Coenye, T. Decreased susceptibility of Streptococcus anginosus to vancomycin in a multispecies biofilm is due to increased thickness of the cell wall. J. Antimicrob. Chemother. 2018, 73, 2323–2330.
- Harriott, M.M.; Noverr, M.C. Candida albicans and Staphylococcus aureus form polymicrobial biofilms: Effects on antimicrobial resistance. Antimicrob. Agents Chemother. 2009, 53, 3914–3922.
- Kean, R.; Rajendran, R.; Haggarty, J.; Townsend, E.M.; Short, B.; Burgess, K.E.; Lang, S.; Millington, O.; Mackay, W.G.; Williams, C.; et al. Candida albicans Mycofilms Support Staphylococcus aureus Colonization and Enhances Miconazole Resistance in Dual-Species Interactions. Front. Microbiol. 2017, 8, 258.
- Kong, E.F.; Tsui, C.; Kucharíková, S.; Van Dijck, P.; Jabra-Rizk, M.A. Modulation of Staphylococcus aureus Response to Antimicrobials by the Candida albicans Quorum Sensing Molecule Farnesol. Antimicrob. Agents Chemother. 2017, 61.
- Flynn, J.M.; Cameron, L.C.; Wiggen, T.D.; Dunitz, J.M.; Harcombe, W.R.; Hunter, R.C. Disruption of Cross-Feeding Inhibits Pathogen Growth in the Sputa of Patients with Cystic Fibrosis. mSphere 2020, 5.
- Hanada, S.; Pirzadeh, M.; Carver, K.Y.; Deng, J.C. Respiratory Viral Infection-Induced Microbiome Alterations and Secondary Bacterial Pneumonia. Front. Immunol. 2018, 9, 2640.
- Bakaletz, L.O. Viral-bacterial co-infections in the respiratory tract. Curr. Opin. Microbiol. 2017, 35, 30–35.
- Bell, L.C.K.; Noursadeghi, M. Pathogenesis of HIV-1 and Mycobacterium tuberculosis co-infection. Nat. Reviews. Microbiol. 2018, 16, 80–90.
- Tonoyan, L.; Vincent-Bugnas, S.; Olivieri, C.V.; Doglio, A. New Viral Facets in Oral Diseases: The EBV Paradox. Int. J. Mol. Sci. 2019, 20, 5861.
- Jossart, G.H.; Canafax, D.M.; Erdmann, G.R.; Lovdahl, M.J.; Russlie, H.Q.; Juhn, S.K.; Giebink, G.S. Effect of Streptococcus pneumoniae and influenza A virus on middle ear antimicrobial pharmacokinetics in experimental otitis media. Pharm. Res. 1994, 11, 860–864.
- Melvin, J.A.; Bomberger, J.M. Compromised Defenses: Exploitation of Epithelial Responses During Viral-Bacterial Co-Infection of the Respiratory Tract. PLoS Pathog. 2016, 12, e1005797.
- Berger, A.K.; Mainou, B.A. Interactions between Enteric Bacteria and Eukaryotic Viruses Impact the Outcome of Infection. Viruses 2018, 10, 19.
- Karst, S.M. The influence of commensal bacteria on infection with enteric viruses. Nat. Reviews. Microbiol. 2016, 14, 197–204.
- Rowe, H.M.; Meliopoulos, V.A.; Iverson, A.; Bomme, P.; Schultz-Cherry, S.; Rosch, J.W. Direct interactions with influenza promote bacterial adherence during respiratory infections. Nat. Microbiol. 2019, 4, 1328–1336.
- Kiedrowski, M.R.; Bomberger, J.M. Viral-Bacterial Co-infections in the Cystic Fibrosis Respiratory Tract. Front. Immunol. 2018, 9, 3067.
