2. High Affinity Phosphate Transport Systems
Most bacteria, including Streptomyces and related actinobacteria, contain high affinity and low affinity phosphate transport systems, which are induced by phosphate limitation, although they may respond to different phosphate thresholds for induction. The wide distribution of both the low affinity and high affinity systems in Gram positive and Gram- negative bacteria indicates that there are physiological advantages in having two different transport systems. The low affinity Pit system requires less energy for phosphate transport since it is energized by a proton-symporter system and is used in high phosphate concentration habitats. The high affinity PstSCAB requires more energy for phosphate transport as it is energized by ATP hydrolysis. This high affinity system is used in habitats or environmental conditions in which phosphate is limiting.
Early evidence of the existence of two phosphate transport systems in Streptomyces was provided by the description of two distinct phosphate uptake kinetics in Streptomyces granaticolor [28], although the genes encoding these systems were not identified.
2.1. The High Affinity PstSCAB System
The high affinity
pstSCAB was first studied in
Escherichia coli [
29],
S. lividans [
20] and
Bacillus subtilis [
30]. In these bacteria, the system consists of four proteins PstS, PstC, PstA, and PstB, but in
B. subtilis there are two copies of PstB (PstB1 and PstB2). The system in
Streptomyces and most other bacteria is organized as an ATP-binding cassette (ABC) that comprises four proteins. Of these, PstS is a phosphate specific binding protein that is anchored in the outer site of the membrane in Gram-positive bacteria, e.g., in
Mycobacterium bovis or
S. lividans [
25,
31,
32], whereas in Gram-negative bacteria, it is located in the periplasmic space. PstC and PstA are membrane integral proteins that form a membrane channel and PstB is an ATP hydrolysing protein that energizes the phosphate transport (). The complete genome sequence of S.
coelicolor [
33] allowed to identify the
pstSCAB operon that is conserved in most
Streptomyces species. Expression of the
pstSCAB cluster responds drastically to the phosphate concentration in the culture medium and is strictly dependent of the activation by the phosphate regulator PhoP [
20,
21,
34]. Deletion of the
pstS gene of
S. lividans or
S. coelicolor impaired phosphate transport and delayed sporulation in solid medium [
34]. In addition to regulation by the PhoP master regulator, formation of the PstS protein was highly increased in a mutant deficient in polyphosphate kinase [
34,
35,
36] which catalyses, in vitro, the reversible polymerization of the phosphate from ATP into polyphosphate. This is most likely due to the up-regulation of
phoR/phoP observed in the
ppk mutant strain [
15]. The formation of PstS increases significantly in media containing fructose, galactose, or mannose, suggesting that control of the
pstSCAB operon is regulated by both inorganic phosphate and some carbon sources.
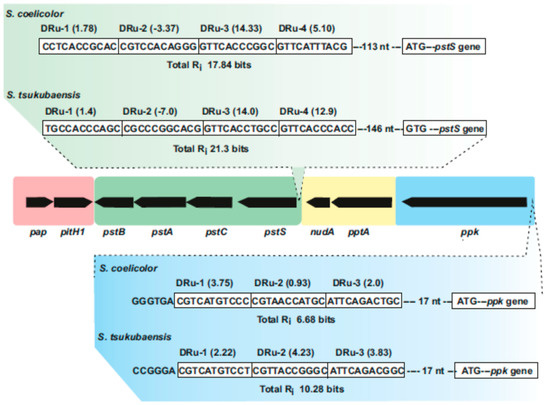
DNA protein binding studies using the purified PhoP protein of
S. coelicolor showed a PHO box consisting of two 11 nucleotides direct repeat units (DRu) upstream of the
pstS gene of
S. coelicolor that adjust to the consensus PhoP-binding sequence [
21,
37]. The
pstSCAB operon of
S. lividans is expressed as a single transcription unit, although levels of the
pstS gene transcript were higher than those of other genes of the operon [
32]. Most likely this is due to the presence of transcription termination sites between the
pstS and
pstC genes. In
S. lividans, deletion of the region encoding the PHO boxes decreased the expression of the
pstSCAB operon, but allows a remaining basal level of expression (about 10%) that was controlled by the carbon source [
32]. The molecular mechanism of regulation by fructose or other carbon sources has not been fully elucidated. CRP (cAMP receptor protein)-binding sequences have been recently identified in the genome of
Streptomyces roseosporus [
38], and a twelve nucleotide repeated sequence upstream of the
pstS gene [
32] might correspond to a CRP binding sequence.
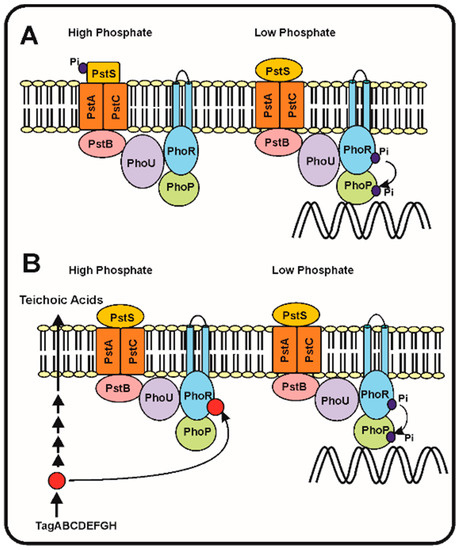
2.2. Sensing the Phosphate Limitation: The Seven Proteins Model of Wanner and the Intracellular Teichoic Acid Intermediate Signal Model
Following the discovery of the two-component system PhoR–PhoB and the
pstSCAB phosphate transport system in
E. coli, models were proposed to try to understand which is the sensing mechanism and, particularly, the transduction cascade that responds to different phosphate concentrations in the periplasmic space. Hsieh and Wanner [
39] elaborated a refined model in which they proposed that seven proteins interact at the membrane level, serving to detect the phosphate concentration in the broth and then transmit the signal to control expression of the Pho regulon genes. This model proposes that the four proteins, PstSCAB, of the ABC transporter system interact with PhoR–PhoB and also with the phosphate modulator PhoU (). According to these authors, depending on the concentration of phosphate in the culture broth, the Pst transporter is either in: (1) an active signalling and transporter state (so called closed state) or, (2) in a state in which it acts as a transporter, but does not transmit signals (open state). The phosphate concentration dependent change between both states produces a rearrangement of the seven proteins complex at the cell membrane, that under phosphate limitation conditions results in autophosphorylation of PhoR, which in turn transfers the phosphate group to PhoB. Finally, the phosphorylated PhoB binds the specific sequences in the promoters of genes of the Pho regulon ().
In the
Streptomyces species, the gene
phoU, encoding a modulator of the
phoR–
phoP expression, is not linked to the
pstSCAB cluster, as occurs in
E. coli, but instead is attached to the
phoR–
phoP cluster [
15,
40]. Signalling of the phosphate concentration and its transport by the PstSCAB cassette in
Streptomyces has received some attention, but is still unknown if the signal interacting with PhoR is internal or extracellular. A second model proposes that, as occurs with
B. subtilis, the signal is an intermediate in the teichoic acid biosynthesis that inhibits the PhoR autokinase activity and, therefore, decreases the phosphorylation of PhoP and the subsequent activation of the Pho regulon [
2,
41]. This correlate well with the presence in
S. coelicolor of a gene cluster for teichoic acid biosynthesis regulated by phosphate similar to that of
B. subtilis [
42]. Under phosphate limiting conditions the phosphorylated teichoic acid intermediate is not accumulated, releasing the inhibition of the PhoR autokinase and the PhoR–PhoP signal transduction cascade proceeds to activate the Pho regulon (B).
Figure 1. Phosphate transport and signalling according to the seven proteins and the teichoic acid intermediate models. (
A) The seven proteins model of Hsieh and Wanner [
39]. (
Left) In phosphate replete conditions, the PstS protein binds phosphate and this interaction blocks the autophosphorylation of PhoR and the subsequent phosphorylation of PhoP avoiding the activation of the genes of the Pho regulon. (
Right). When phosphate is limiting the PstS conformation changes, and triggers the phosphorylation cascade, resulting in PhoP phosphorylation and activation of the Pho regulon. (
B) Proposed model of regulation of the phosphorylation cascade by an intermediate of the teichoic acid biosynthesis (red circle). Under phosphate replete conditions (left side) the teichoic acid intermediate accumulates and inhibits the autokinase activity of PhoR, blocking the phosphorylation cascade. Under phosphate limitation conditions (right side), the teichoic acid intermediate is not accumulated [
41] and, therefore, the phosphorylation cascade proceeds activating expression of the Pho regulon genes (see text for details).
Figure 2. Supercluster of genes involved in phosphate uptake and metabolism in Streptomyces species. The central region includes the pap–pitH1 operon (shadowed in pink), the pstSCAB cluster (green), the pptA–nudA genes (yellow) and the ppk gene (blue). In the upper part, the PHO boxes upstream of pstS in S. coelicolor and Streptomyces tsukubaensis are shown indicating the direct repeat units (Dru’s) (boxed) and their individual information values (Ri). In the lower part updated PHO boxes upstream of the ppk gene in S. coelicolor and S. tsukubaensis are shown. Note that the pap–pitH1 operon is not regulated by phosphate and does not contain PHO boxes.
A third hypothesis is also possible: PhoU and the metalloprotein encoded by
mtpA, a gene adjacent to
phoU, might be involved in the sensing or resistance to oxidative stress [
43,
44]. Since PhoU it is known to modulate expression of the
phoR–phoP operon [
15,
40], it is possible that oxidative stress might lead to a modified PhoU conformation conferring to PhoU the ability to stimulate the PhoR autophosphorylation activity. Studies on the conformational changes of PhoU are required to support this hypothesis.
2.3. Discrimination between the Phosphate and Arsenate Anions by the Phosphate-Binding Protein PstS
Phosphate (PO
43−) and arsenate (AsO
43−) are equivalent anions with similar pK(a) values and charges in their atoms. Arsenate, a toxic anion, is present in some terrestrial and aquatic habitats and, therefore,
Streptomyces and other bacteria face the problem of discriminating between phosphate and arsenate. In previous studies, we observed in the polyene macrolide antibiotic candicidin producer
Streptomyces griseus, that arsenate is toxic at concentrations above 10 mM. Expression of the genes for candicidin biosynthesis and candicidin formation are strictly regulated by the phosphate concentration in the cultures [
45,
46]. Some mutants resistant to arsenate (up to 100 mM) are impaired in the phosphate transport, as determined by radioactive phosphate uptake experiments, whereas others showed normal phosphate uptake [
47]. This suggests that somehow there is a competition between phosphate and arsenate, either in the phosphate transport system or perhaps downstream in the signal transducing mechanisms. Several mutants resistant to arsenate overproduced candicidin at phosphate levels (10 mM) that were repressive for candicidin biosynthesis in the parental strain. The molecular mechanism responsible of the candicidin overproduction have not been studied so far.
The PstS proteins have a great affinity for phosphate with Kd values in the submicromolar range. The affinity of the PstS protein for phosphate is 500 to 700 higher than that for arsenate [
48]; even more, the PstS protein of a
Halomonas strain isolated from a arsenate rich environment has 4500-fold higher affinity for phosphate than for arsenate. Crystal structure of several bacterial PstS proteins [
27,
49] indicates that they are composed by two globular domains linked together by a flexible hinge. At the interface between the domains, there is a phosphate binding pocket containing a key aspartate residue. Twelve to fourteen hydrogen bonds are formed between the phosphate and the binding pocket, and the aspartate residue establishes an additional low barrier hydrogen bond. The presence of both, high and low hydrogen barrier bonds, confers high selectivity for phosphate. A PstS protein with higher affinity for arsenate has been crystalized from
Clostridium perfringens; this protein still forms 14 hydrogen bonds, but lacks the low barrier bond between the phosphate and the aspartate residue [
49]. The arsenate anion is 4% larger than the phosphate anion; its accommodation in PstS requires the distortion of this low barrier hydrogen bond allowing its discrimination by distorting this low barrier hydrogen bond. No PstS proteins of
Streptomyces have been yet crystalized to confirm whether similar mechanism occurs in these bacteria.
2.4. Glycosylation and Release of PstS in Streptomyces Species: Which Is the Role of the Released PstS Protein?
The PstS proteins are usually cell membrane-anchored proteins in different bacteria. However, surprisingly it was reported that in some Streptomyces species this protein was released to the culture medium.
The PstS protein of
S. coelicolor is glycosylated [
31]. Glycosylation in actinobacteria has been well studied in
M. tuberculosis and proceeds through O-mannosylation of threonine or serine residues located in a proline rich region of the target protein. Several extracellular antigens are glycosylated in
M. tuberculosis, including the 45/47 kDa, the 19 kDa and the main 38 kDa antigen protein, identified as PstS1 [
23,
24]. The carbohydrate moieties include mannose, mannobiose and mannotriose [
24]. The glycosylation proceeds first through activation of mannose with polyprenol phosphate by a polyprenol phosphate mannose synthase (Pmm). A second gene involved in the glycosylation process is
pmt1 that encodes a polyprenol mannose transferase.
S. lividans contains homologous genes encoding the same enzymes than
M. tuberculosis. The first evidence of the involvement of these two enzymes in glycosylation in
Streptomyces came from studies of the glycosylation of two phage receptor proteins in
S. coelicolor [
50,
51]. The enzymes encoded by the
pmm and
pmt1 genes of
S. lividans were able to glycosylate antigens and extracellular enzymes of
M. tuberculosis [
52,
53] confirming the similarity of the glycosylation systems in both actinobacteria. In addition to its role in the synthesis of glycoproteins that serve as receptor of phages in
S. coelicolor, Wehmeier et al. [
31] found that these two enzymes were also involved in the glycosylation of the PstS phosphate binding protein. In
S. coelicolor glycosylation involves the introduction of three hexose molecules and does not occur in
pmt1 or
pmm impaired mutants [
31]. The two enzymes were functional to glycosylate PstS peptides bound to membranes of
S. coelicolor. The physiological role of the PstS modification by glycosylation is unclear. Glycosylation of proteins in prokaryotic and eukaryotic cells is connected with protein sorting, protein–protein interactions and membrane localization.
The PstS protein of
S. lividans is released from the mycelium to the culture broth, and interestingly the free form is unable to bind phosphate at difference of the cell-anchored PstS protein. In summary, expression of the
pstS gene in
S. coelicolor drastically increases in response to phosphate starvation and, therefore, the protein is accumulated in the cell membrane and under adequate conditions is released. In some bacteria, it has been reported that the PstS protein has additional roles different from serving as the phosphate receptor protein. For example, in multi-drug resistant
Pseudomonas aeruginosa the extracellular PstS forms appendage-like structures that interact with human epithelial intestinal cells disturbing their permeability [
54]. It is then intriguing to know if the release of the PstS protein of
S. coelicolor may have functions resulting in interaction and communication of the producer
Streptomyces species with other soil microorganisms or with animal tissues in the case of pathogenic
Streptomyces species, although there is no evidence so far supporting this hypothesis.
2.5. A Second High Affinity Phosphate Transport System Exists in Mycobacterium smegmatis
The slow growing actinobacteria M. tuberculosis contain several high affinity transport systems similar to pstSCAB (two to four copies) whereas fast growing mycobacteria contain usually a single copy of the pstSCAB cluster. The PstS1, Pst2, and PstS3 phosphate sensor proteins of M. tuberculosis have been studied in detail [26] and some of them have been crystalized [27]. The fast growing Mycobacterium smegmatis contains two high affinity phosphate transport systems of different types; in addition to the classical pstSCAB phosphate transport system, a second transport system named phnFDCE (so named because it was initially proposed to be a phosphonate transporter) was characterized [55,56]. The PhnDCE transport system, an ABC-type transporter, consists in three structural proteins: phnC encodes an ATPase, phnD a phosphate-binding protein and phnE encodes a permease. The phnF gene, transcribed in opposite orientation encodes a regulator of the GntR family that controls the phnCDE cluster [57]. The PhnF protein represses expression of the phnDCE cluster by binding to two nucleotide sequences in the bidirectional promoter regions [57]. This system transports Pi with relatively high affinity (Km values of 40 to 90 micromolar), and does not transport phosphonate or phosphite, a salt of phosphorous acid [58]. Expression of the phnDCE cluster increased under phosphate limitation conditions. Mutants defective in the phosphate-binding protein PhnD, failed to grow in minimal medium containing at high phosphate concentrations (10 mM) while the parental strain requires only micromolar phosphate concentrations. Complementation of the null phnD mutant with the wild type gene resulted in recovery of the growth of this strain at submicromolar phosphate concentrations [55]. In summary, the available evidence indicates that the M. smegmatis phnFDCE transport system is a second high affinity transporter for inorganic phosphate at difference of the Phn system in Gram negative bacteria that has been reported to transport both phosphonate and inorganic phosphate [59,60,61,62]. Gene clusters similar to phnCDE exist in many Streptomyces species, but only occasionally they are linked to the phosphonate transport and utilization clusters.