Solution nuclear magnetic resonance (NMR) spectroscopy is a promising tool in drug discovery. Especially, fragment-based drug discovery (FBDD) has benefited a lot from the NMR development. Multiple candidate compounds and FDA-approved drugs derived from FBDD have been developed with the assistance of NMR techniques. NMR has broad applications in different stages of the FBDD process, which includes fragment library construction, hit generation and validation, hit-to-lead optimization and working mechanism elucidation, etc.
- nuclear magnetic resonance (NMR)
- Drug Discovery
1. Introduction
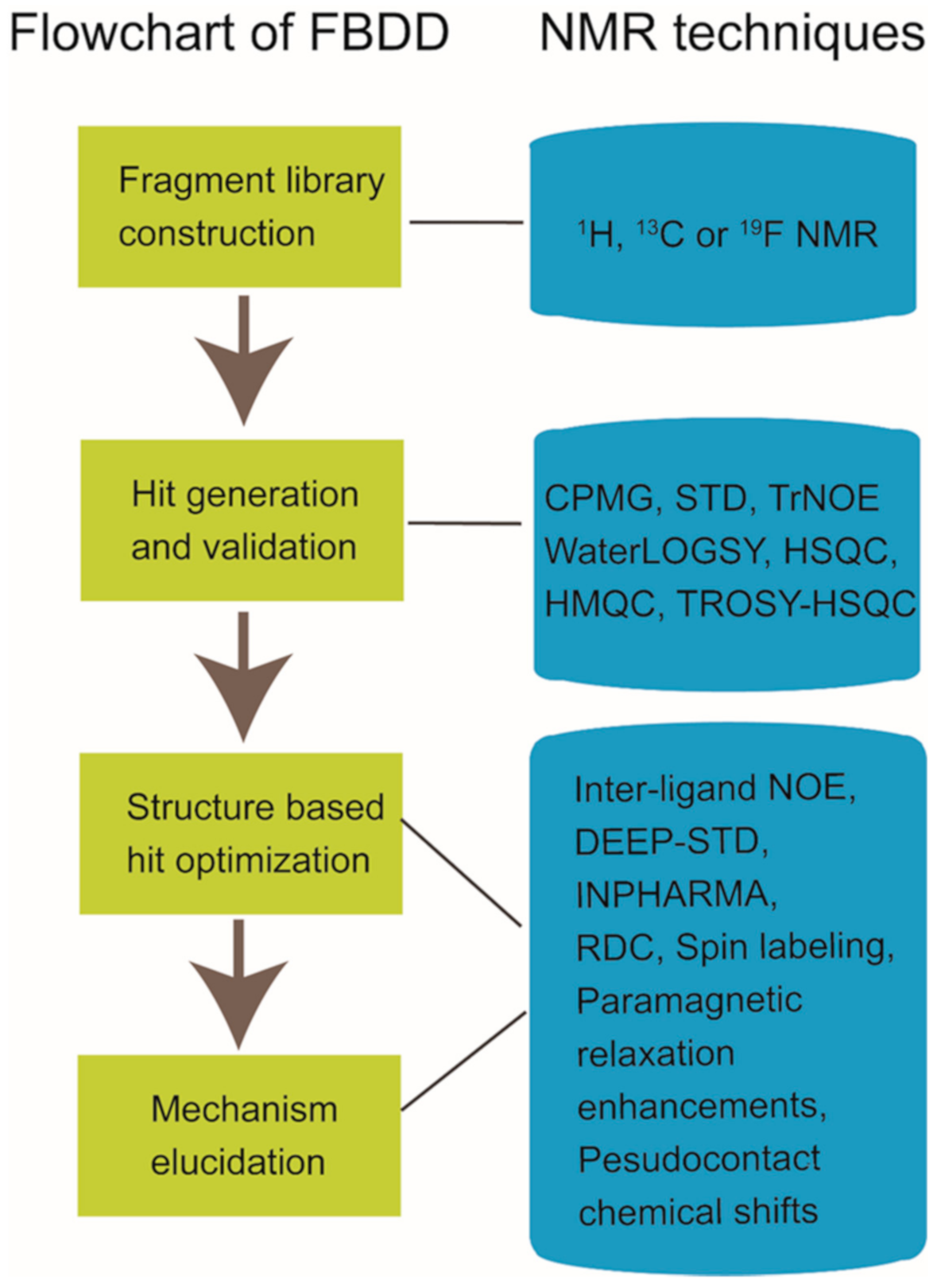
Nuclear magnetic resonance (NMR) spectroscopy has been widely used in structure determination and dynamics investigation of biomacromolecules under physiological conditions. Meanwhile, due to its advantages in detecting transient and weak interactions, NMR has been becoming a powerful tool in drug discovery [1][2][3][4]. FBDD (fragment-based drug discovery), which serves as a key approach for finding high-quality lead candidates, has benefited a lot from NMR spectroscopy development [5][6][7][8][9][10]. Accumulated studies have shown the extensive applications of solution NMR in FBDD field (Figure 1), which include fragment library construction, ligand-observed and target-observed hit screening and validation, etc. [2][11]. During the past decade, FBDD has established itself as a promising drug discovery approach, which has been applied in candidate compound developing for various drug targets such as DNA, RNA, kinases, enzymes, membrane proteins, and even inherently disordered proteins [12][13][14][15]. Fragment compounds for FBDD are small organic molecules with their molecular weights typically not exceeding 300 Da, and due to the limited molecular size of fragment compounds, their binding affinities to the targets usually fall into the micromolar to millimolar range [16]. NMR spectroscopy, which is sensitive to weak interactions, is one of the top choices for hit compound screening against a fragment compound library [17]. Meanwhile, since target-observed NMR techniques are capable of providing structural information for structure-guided hit fragment optimization [18][19], they are alternative methods to X-ray crystallography for the characterization of target-hit/lead interaction. Dynamics investigation, inter-molecular NOEs (nuclear Overhauser effects) and paramagnetic NMR can help to reveal atomic level details associated with the binding mode of the hit or lead to defined target [20][21][22][23][24]. Although NMR data collection and data processing is time-consuming, this is especially necessary when researchers devote great efforts to obtain the crystal structures of target-hit/lead complexes but in vain [25].
Except for the conventional target-oriented drug discovery, targeting protein-protein interactions (PPIs) has been emerging as an attractive approach for drug development. The protein-protein interaction network, the so-called interactome, participates in extensive biological processes, and aberrant expression or regulation of the interactome would cause the occurrence of severe human diseases [26][27][28][29]. PPI modulators, which present increased target or signal pathway selectivity and decreased off-target side effects, have high potentials for therapeutic uses [30]. Hence, the discovery and development of chemical compounds to modulate the interactome has gained enormous attention. Compared with the typical binding pockets for small molecules in conventional targets, the surface areas for protein-protein interactions are often large and flat, which introduces more challenges to the PPI-targeted drug development [31][32][33]. However, it is worth mentioning that NMR and FBDD have been becoming powerful tools in developing drugs for the ‘’undruggable’’ targets due to their advantages in dynamic and transient systems such as protein-protein interactome [26][34][35][36]. Multiple PPI-targeted hit compounds against fragment compound libraries have been developed [37][38][39][40][41], and a few of PPI stabilizers and breakers have been validated by NMR [38][39][42]. GNE667, a novel inhibitor of deubiquitinase USP7, was found to disrupt the interaction between USP7 and its native substrate ubiquitin [38]. CC0651 inhibited the activity of Cdc34A (an E2 enzyme) by enhancing the binding affinity of ubiquitin to Cdc34A, thus blocking the discharging process of ubiquitin to E3 ligase [39].
Similar to the in vitro NMR applications in drug development, NMR techniques could also be conducted to detect target-compound interactions in living cells such as E.coli, yeast and mammalian cells [43][44]. Researchers have applied in-cell NMR techniques in compound screening and target engagement, which are two important aspects of drug discovery [43][45]. However, there are still several factors need to be considered as challenges remaining in the in-cell NMR studies, which include molecular size limitation, high background signals, nonspecific interactions in cells and protein leakage produced by dead cells [44][46][47][48][49].
2. Applications of Solution NMR in Drug Discovery
Solution NMR spectroscopy is a well-established approach to elucidate the structure, interaction, and dynamics of molecules in physiological conditions, and it has become a powerful tool in drug discovery. In fact, over the past decades, NMR has been widely used in drug-related research, especially in fragment-based drug discovery. NMR has a broader application in supporting FBDD, which is capable in fragment library construction, hit fragment screening, and binding mode characterization for the guidance of structure-based optimization [6][8][18][50]. To extend the application scope of NMR in drug discovery, scientists have devoted great efforts into the field. Isotope labeling, non-uniform sampling, reduced dimensionality techniques for rapid measurements, and automated software for NMR data analysis have been developed to improve the efficiency of NMR experiments [51][52][53][54][55]. Different NMR techniques such as selective paramagnetic labeling of target or ligand, INPHARMA, etc. have also been tried in exploring the structural information of compound/target complexes [56][57][58].
NMR spectroscopy has also demonstrated itself as a powerful tool in PPI modulator development. Drug development targeting protein-protein interactions has long been considered as a very difficult and even impossible task. Designing peptides mimicking amino acid residues in the PPI interface is a rational starting point to design PPI modulators; however, the bioavailability and the in vivo stability of these peptides is usually very low [29]. Therefore, small molecules targeting the PPI network have caught people’s eyes [36]. Different from the druggable pockets in conventional drug targets, the protein-protein binding surface is usually flat and undruggable. Fragmentation and NMR are robust tools for developing PPI modulators. As a convincing example, NMR-based fragment hit screening and the characterization of hit-target interactions contributed significantly to the successful development of the FDA-approved drug Venetoclax (ABT-199).
In addition to the in vitro applications, NMR spectroscopy could also be applied to support drug discovery in a cellular context. In-cell NMR has been developed for nearly 20 years. Researchers have applied the in-cell NMR method to hit compound screening and interaction characterization of the compound/target system in different types of living cells including prokaryotes cells and human cancer cells [43][59]. Of note, severe line broadening of NMR spectra caused by the crowded cellular environment restricts the application of in-cell NMR [60]. New strategies including selective isotope labeling and high-resolution magic angle spinning (HR-MAS) etc. have been developed to improve the quality of in-cell NMR spectra [61][62]. For example, due to the low abundance in biomacromolecules and the high sensitivity, 19F has been utilized as an important probe to investigate target-ligand interactions in living cells [63][64]. Cell death is another issue that needs to be addressed in conducting in-cell NMR. Fast pulse sequences and various bioreactors were developed to shorten in-cell NMR data collection time and improve cell viability [65][66][67].
NMR, X-ray, and cryo-EM are three major structural biology techniques that have been widely used in industry and academia. Compared to NMR, X-ray and cryo-EM exhibit robust advantages on large complex systems or biomolecule machineries. However, although NMR has the limitation for biomacromolecules with large molecular weights, NMR spectroscopy presents particular merits in detecting the structural information of dynamic biomacromolecule systems. In addition, NMR is one of the most promising techniques in studying target-ligand interactions in living cells, which is pretty important for drug evaluation.
This entry is adapted from the peer-reviewed paper 10.3390/molecules26030576
References
- Emwas, A.H.; Szczepski, K.; Poulson, B.G.; Chandra, K.; McKay, R.T.; Dhahri, M.; Alahmari, F.; Jaremko, L.; Lachowicz, J.I.; Jaremko, M. NMR as a “Gold Standard” Method in Drug Design and Discovery. Molecules 2020, 25, 4597.
- Li, Q.; Kang, C.B. A Practical Perspective on the Roles of Solution NMR Spectroscopy in Drug Discovery. Molecules 2020, 25, 2974.
- Pellecchia, M.; Bertini, I.; Cowburn, D.; Dalvit, C.; Giralt, E.; Jahnke, W.; James, T.L.; Homans, S.W.; Kessler, H.; Luchinat, C.; et al. Perspectives on NMR in drug discovery: A technique comes of age. Nat. Rev. Drug Discov. 2008, 7, 738–745.
- Shuker, S.B.; Hajduk, P.J.; Meadows, R.P.; Fesik, S.W. Discovering high-affinity ligands for proteins: SAR by NMR. Science 1996, 274, 1531–1534.
- Lamoree, B.; Hubbard, R.E. Current perspectives in fragment-based lead discovery (FBLD). Essays Biochem. 2017, 61, 453–464.
- Harner, M.J.; Frank, A.O.; Fesik, S.W. Fragment-based drug discovery using NMR spectroscopy. J. Biomol. NMR 2013, 56, 65–75.
- Li, Q. Application of Fragment-Based Drug Discovery to Versatile Targets. Front. Mol. Biosci. 2020, 7, 180.
- Murray, C.W.; Rees, D.C. The rise of fragment-based drug discovery. Nat. Chem. 2009, 1, 187–192.
- Ayotte, Y.; Murugesan, J.R.; Bilodeau, F.; Larda, S.; Bouchard, P.; Drouin, N.; Morin, M.; LaPlante, S. Discovering Quality Drug Seeds by Practical NMR-based Fragment Screening. Protein Sci. 2017, 26, 194–195.
- Erlanson, D.A.; Fesik, S.W.; Hubbard, R.E.; Jahnke, W.; Jhoti, H. Twenty years on: The impact of fragments on drug discovery. Nat. Rev. Drug Discov. 2016, 15, 605–619.
- Ma, R.; Wang, P.; Wu, J.; Ruan, K. Process of Fragment-Based Lead Discovery-A Perspective from NMR. Molecules 2016, 21, 854.
- Yanamala, N.; Dutta, A.; Beck, B.; van Fleet, B.; Hay, K.; Yazbak, A.; Ishima, R.; Doemling, A.; Klein-Seetharaman, J. NMR-based screening of membrane protein ligands. Chem. Biol. Drug Des. 2010, 75, 237–256.
- Iconaru, L.I.; Ban, D.; Bharatham, K.; Ramanathan, A.; Zhang, W.; Shelat, A.A.; Zuo, J.; Kriwacki, R.W. Discovery of Small Molecules that Inhibit the Disordered Protein, p27(Kip1). Sci. Rep. 2015, 5, 15686.
- Han, B.; Ahn, H.C. Recombinant Kinase Production and Fragment Screening by NMR Spectroscopy. Methods Mol. Biol. 2016, 1360, 35–46.
- Binas, O.; de Jesus, V.; Landgraf, T.; Volklein, A.E.; Martins, J.; Hymon, D.; Bains, J.K.; Berg, H.; Biedenbander, T.; Furtig, B.; et al. F-19 NMR-Based Fragment Screening for 14 Different Biologically Active RNAs and 10 DNA and Protein Counter-Screens. ChemBioChem 2020, 22, 423–433.
- Keseru, G.M.; Erlanson, D.A.; Ferenczy, G.G.; Hann, M.M.; Murray, C.W.; Pickett, S.D. Design Principles for Fragment Libraries: Maximizing the Value of Learnings from Pharma Fragment-Based Drug Discovery (FBDD) Programs for Use in Academia. J. Med. Chem. 2016, 59, 8189–8206.
- Kobayashi, M.; Retra, K.; Figaroa, F.; Hollander, J.G.; Ab, E.; Heetebrij, R.J.; Irth, H.; Siegal, G. Target Immobilization as a Strategy for NMR-Based Fragment Screening: Comparison of TINS, STD, and SPR for Fragment Hit Identification. J. Biomol. Screen. 2010, 15, 978–989.
- Huth, J.R.; Sun, C. Utility of NMR in lead optimization: Fragment-based approaches. Comb. Chem. High Throughput Screen. 2002, 5, 631–643.
- Recht, M.I.; Sridhar, V.; Badger, J.; Bounaud, P.Y.; Logan, C.; Chie-Leon, B.; Nienaber, V.; Torres, F.E. Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography. J. Biomol. Screen. 2014, 19, 497–507.
- Stark, J.L.; Powers, R. Application of NMR and molecular docking in structure-based drug discovery. Top. Curr. Chem. 2012, 326, 1–34.
- Orts, J.; Gossert, A.D. Structure determination of protein-ligand complexes by NMR in solution. Methods 2018, 138, 3–25.
- Sugiki, T.; Furuita, K.; Fujiwara, T.; Kojima, C. Current NMR Techniques for Structure-Based Drug Discovery. Molecules 2018, 23, 148.
- Bernini, A.; Henrici De Angelis, L.; Morandi, E.; Spiga, O.; Santucci, A.; Assfalg, M.; Molinari, H.; Pillozzi, S.; Arcangeli, A.; Niccolai, N. Searching for protein binding sites from Molecular Dynamics simulations and paramagnetic fragment-based NMR studies. Biochim. Biophys. Acta 2014, 1844, 561–566.
- Softley, C.A.; Bostock, M.J.; Popowicz, G.M.; Sattler, M. Paramagnetic NMR in drug discovery. J. Biomol. NMR 2020, 74, 287–309.
- Erlanson, D.A.; Davis, B.J.; Jahnke, W. Fragment-Based Drug Discovery: Advancing Fragments in the Absence of Crystal Structures. Cell Chem. Biol. 2019, 26, 9–15.
- Dias, D.M.; Van Molle, I.; Baud, M.G.; Galdeano, C.; Geraldes, C.F.; Ciulli, A. Is NMR Fragment Screening Fine-Tuned to Assess Druggability of Protein-Protein Interactions? ACS Med. Chem. Lett. 2014, 5, 23–28.
- Valkov, E.; Sharpe, T.; Marsh, M.; Greive, S.; Hyvonen, M. Targeting protein-protein interactions and fragment-based drug discovery. Top. Curr. Chem. 2012, 317, 145–179.
- Bosch, J. PPI inhibitor and stabilizer development in human diseases. Drug Discov. Today Technol. 2017, 24, 3–9.
- Lu, H.Y.; Zhou, Q.D.; He, J.; Jiang, Z.L.; Peng, C.; Tong, R.S.; Shi, J.Y. Recent advances in the development of protein-protein interactions modulators: Mechanisms and clinical trials. Signal Transduct. Target. Ther. 2020, 5, 1–23.
- Wang, L.; Zhang, L.; Li, L.; Jiang, J.; Zheng, Z.; Shang, J.; Wang, C.; Chen, W.; Bao, Q.; Xu, X.; et al. Small-molecule inhibitor targeting the Hsp90-Cdc37 protein-protein interaction in colorectal cancer. Sci. Adv. 2019, 5, 1–13.
- Arkin, M.R.; Tang, Y.; Wells, J.A. Small-molecule inhibitors of protein-protein interactions: Progressing toward the reality. Chem. Biol. 2014, 21, 1102–1114.
- Mabonga, L.; Kappo, A.P. Protein-protein interaction modulators: Advances, successes and remaining challenges. Biophys. Rev. 2019, 11, 559–581.
- Ni, D.; Lu, S.Y.; Zhang, J. Emerging roles of allosteric modulators in the regulation of protein-protein interactions (PPIs): A new paradigm for PPI drug discovery. Med. Res. Rev. 2019, 39, 2314–2342.
- Brancaccio, D.; Di Maro, S.; Cerofolini, L.; Giuntini, S.; Fragai, M.; Luchinat, C.; Tomassi, S.; Limatola, A.; Russomanno, P.; Merlino, F.; et al. HOPPI-NMR: Hot-Peptide-Based Screening Assay for Inhibitors of Protein-Protein Interactions by NMR. ACS Med. Chem. Lett. 2020, 11, 1047–1053.
- Sijbesma, E.; Hallenbeck, K.K.; Leysen, S.; de Vink, P.J.; Skora, L.; Jahnke, W.; Brunsyeld, L.; Arkin, M.R.; Ottmann, C. Site-Directed Fragment-Based Screening for the Discovery of Protein-Protein Interaction Stabilizers. J. Am. Chem. Soc. 2019, 141, 3524–3531.
- Sheng, C.Q.; Dong, G.Q.; Miao, Z.Y.; Zhang, W.N.; Wang, W. State-of-the-art strategies for targeting protein-protein interactions by small-molecule inhibitors (vol 44, pg 8238, 2015). Chem. Soc. Rev. 2015, 44, 8238–8259.
- Chen, D.; Chen, Y.; Lian, F.; Chen, L.; Li, Y.; Cao, D.; Wang, X.; Chen, L.; Li, J.; Meng, T.; et al. Fragment-based drug discovery of triazole inhibitors to block PDEdelta-RAS protein-protein interaction. Eur. J. Med Chem. 2019, 163, 597–609.
- Kategaya, L.; Di Lello, P.; Rouge, L.; Pastor, R.; Clark, K.R.; Drummond, J.; Kleinheinz, T.; Lin, E.; Upton, J.P.; Prakash, S.; et al. USP7 small-molecule inhibitors interfere with ubiquitin binding. Nature 2017, 550, 534–538.
- Huang, H.; Ceccarelli, D.F.; Orlicky, S.; St-Cyr, D.J.; Ziemba, A.; Garg, P.; Plamondon, S.; Auer, M.; Sidhu, S.; Marinier, A.; et al. E2 enzyme inhibition by stabilization of a low-affinity interface with ubiquitin. Nat. Chem. Biol. 2014, 10, 156–163.
- Milroy, L.G.; Bartel, M.; Henen, M.A.; Leysen, S.; Adriaans, J.M.; Brunsveld, L.; Landrieu, I.; Ottmann, C. Stabilizer-Guided Inhibition of Protein-Protein Interactions. Angew. Chem. Int. Ed. Engl. 2015, 54, 15720–15724.
- Bower, J.F.; Pannifer, A. Using fragment-based technologies to target protein-protein interactions. Curr. Pharm. Des. 2012, 18, 4685–4696.
- Moriya, J.; Takeuchi, K.; Tai, K.J.; Arai, K.; Kobayashi, N.; Yoneda, N.; Fukunishi, Y.; Inoue, A.; Kihara, M.; Murakami, T.; et al. Structure-Based Development of a Protein-Protein Interaction Inhibitor Targeting Tumor Necrosis Factor Receptor-Associated Factor 6. J. Med. Chem. 2015, 58, 5674–5683.
- Kang, C. Applications of In-Cell NMR in Structural Biology and Drug Discovery. Int. J. Mol. Sci. 2019, 20, 139.
- Siegal, G.; Selenko, P. Cells, drugs and NMR. J. Magn. Reson. 2019, 306, 202–212.
- Cerofolini, L.; Giuntini, S.; Barbieri, L.; Pennestri, M.; Codina, A.; Fragai, M.; Banci, L.; Luchinat, E.; Ravera, E. Real-Time Insights into Biological Events: In-Cell Processes and Protein-Ligand Interactions. Biophys. J. 2019, 116, 239–247.
- Maldonado, A.Y.; Burz, D.S.; Shekhtman, A. In-cell NMR spectroscopy. Prog. Nucl. Magn. Reson. Spectrosc. 2011, 59, 197–212.
- Luchinat, E.; Banci, L. In-Cell NMR in Human Cells: Direct Protein Expression Allows Structural Studies of Protein Folding and Maturation. Acc. Chem. Res. 2018, 51, 1550–1557.
- Selenko, P.; Wagner, G. Looking into live cells with in-cell NMR spectroscopy. J. Struct. Biol. 2007, 158, 244–253.
- Li, C.; Zhao, J.; Cheng, K.; Ge, Y.; Wu, Q.; Ye, Y.; Xu, G.; Zhang, Z.; Zheng, W.; Zhang, X.; et al. Magnetic Resonance Spectroscopy as a Tool for Assessing Macromolecular Structure and Function in Living Cells. Annu. Rev. Anal. Chem. 2017, 10, 157–182.
- Campos-Olivas, R. NMR screening and hit validation in fragment based drug discovery. Curr. Top. Med. Chem. 2011, 11, 43–67.
- Takeuchi, K.; Arthanari, H.; Shimada, I.; Wagner, G. Nitrogen detected TROSY at high field yields high resolution and sensitivity for protein NMR. J. Biomol. Nmr 2015, 63, 323–331.
- Orts, J.; Riek, R. Protein-ligand structure determination with the NMR molecular replacement tool, NMR(2). J. Biomol. NMR 2020, 74, 633–642.
- Fino, R.; Byrne, R.; Softley, C.A.; Sattler, M.; Schneider, G.; Popowicz, G.M. Introducing the CSP Analyzer: A novel Machine Learning-based application for automated analysis of two-dimensional NMR spectra in NMR fragment-based screening. Comput. Struct. Biotechnol. J. 2020, 18, 603–611.
- Strotz, D.; Orts, J.; Chi, C.N.; Riek, R.; Vogeli, B. eNORA2 Exact NOE Analysis Program. J. Chem. Theory Comput. 2017, 13, 4336–4346.
- Reynolds, W.F.; Breton, R.C.; Burns, D.C. Evaluating ASAP-HMQC and PS-HSQC NMR pulse sequences with non-uniform sampling for rapid screening of natural products. Planta Med. 2014, 80, IL26.
- Codutti, L.; Grimaldi, M.; Carlomagno, T. Structure-Based Design of Scaffolds Targeting PDE10A by INPHARMA-NMR. J. Chem. Inf. Modeling 2017, 57, 1488–1498.
- Sikorska, J.; Codutti, L.; Ameneiro, R.S.; Skjaerven, L.; Angelini, A.; Carlomagno, T. Understanding of Protein-Ligand Interactions with INPHARMA. Planta Med. 2013, 79, PD2.
- Mateos, B.; Konrat, R.; Pierattelli, R.; Felli, I.C. NMR Characterization of Long-Range Contacts in Intrinsically Disordered Proteins from Paramagnetic Relaxation Enhancement in C-13 Direct-Detection Experiments. ChemBioChem 2019, 20, 335–339.
- Nishida, N.; Ito, Y.; Shimada, I. In situ structural biology using in-cell NMR. Biochim. Biophysca Acta Gen. Subj. 2020, 1864, 129364.
- Luchinat, E.; Banci, L. In-cell NMR: A topical review. IUCrJ 2017, 4, 108–118.
- Tripsianes, K.; Schutz, U.; Emmanouilidis, L.; Gemmecker, G.; Sattler, M. Selective isotope labeling for NMR structure determination of proteins in complex with unlabeled ligands. J. Biomol. Nmr 2019, 73, 183–189.
- Airoldi, C.; Giovannardi, S.; La Ferla, B.; Jimenez-Barbero, J.; Nicotra, F. Saturation transfer difference NMR experiments of membrane proteins in living cells under HR-MAS conditions: The interaction of the SGLT1 co-transporter with its ligands. Chemistry 2011, 17, 13395–13399.
- Ye, Y.S.; Liu, X.L.; Zhang, Z.T.; Wu, Q.; Jiang, B.; Jiang, L.; Zhang, X.; Liu, M.L.; Pielak, G.J.; Li, C.G. F-19 NMR Spectroscopy as a Probe of Cytoplasmic Viscosity and Weak Protein Interactions in Living Cells. Chem. Eur. J. 2013, 19, 12705–12710.
- Li, C.G.; Wang, G.F.; Wang, Y.Q.; Creager-Allen, R.; Lutz, E.A.; Scronce, H.; Slade, K.M.; Ruf, R.A.S.; Mehl, R.A.; Pielak, G.J. Protein F-19 NMR in Escherichia coli. J. Am. Chem. Soc. 2010, 132, 321–327.
- Kubo, S.; Nishida, N.; Udagawa, Y.; Takarada, O.; Ogino, S.; Shimada, I. A Gel-Encapsulated Bioreactor System for NMR Studies of Protein-Protein Interactions in Living Mammalian Cells. Angew. Chem. Int. Ed. 2013, 52, 1208–1211.
- Sharaf, N.G.; Barnes, C.O.; Charlton, L.M.; Young, G.B.; Pielak, G.J. A bioreactor for in-cell protein NMR. J. Magn. Reson. 2010, 202, 140–146.
- Motta, A.; Paris, D.; Melck, D. Monitoring Real-Time Metabolism of Living Cells by Fast Two-Dimensional NMR Spectroscopy. Anal. Chem. 2010, 82, 2405–2411.
