The interaction between the plant host, walnut (Juglans regia; Jr), and a deadly pathogen (Xanthomonas arboricola pv. juglandis 417; Xaj) can lead to walnut bacterial blight (WB), which depletes walnut productivity by degrading the nut quality. Here, we dissect this pathosystem using tandem mass tag quantitative proteomics. Walnut hull tissues inoculated with Xaj were compared to mock-inoculated tissues, and 3972 proteins were identified, of which 3296 are from Jr and 676 from Xaj. Proteins with differential abundance include oxidoreductases, proteases, and enzymes involved in energy metabolism and amino acid interconversion pathways. Defense responses and plant hormone biosynthesis were also increased. Xaj proteins detected in infected tissues demonstrate its ability to adapt to the host microenvironment, limiting iron availability, coping with copper toxicity, and maintaining energy and intermediary metabolism. Secreted proteases and extracellular secretion apparatus such as type IV pilus for twitching motility and type III secretion effectors indicate putative factors recognized by the host. Taken together, these results suggest intense degradation processes, oxidative stress, and general arrest of the biosynthetic metabolism in infected nuts. Our results provide insights into molecular mechanisms and highlight potential molecular tools for early detection and disease control strategies.
- Xanthomonas
- walnut blight
- fruit
- proteomics
- disease susceptibility
- adaptation
- LC-MS/MS
- virulence
1. Introduction
Copper-based biocides are effective in many pathosystems involving Xanthomonas spp., although some Xaj isolates are especially resistant [1][2]. Some pathosystems such as the bacterial blight of rice, bacterial spot, and black rot diseases have been studied in molecular detail, revealing specific plant-microbe protein interactions [3][4][5][6][7]. These include, for example, production by the host of pathogenesis-related (PR) proteins triggered by detection of pathogen- or damage-associated molecular patterns (PAMPs and DAMPs, respectively) [8][9]. Another layer of responses involves effector-triggered immunity (ETI) upon the detection of type III effectors secreted into plant cells by the bacterium, capable of altering the host gene expression and disease progression [10]. Similar strategies are also employed by Xanthomonas pathogens in stone and fruit trees, exemplified by citrus bacterial canker disease caused by Xanthomonas citri [11]. Despite these similarities, there are also significant differences such as the adoption of transcription activator-like (TAL) effectors to alter host responses by some, but not all, Xanthomonas species [12]. Hence, the need to study each pathosystem in molecular detail, so common and disease-specific themes can be identified and characterized more precisely. In WB, a distinctive symptom on fruits is the intensely dark pigmentation of the hull tissues that progresses to the edible parts, until the kernel is damaged [13]. Polyphenol oxidases (PPOs), which are copper-containing enzymes, have been implicated in the synthesis of these pigments, which include tannins and melanins [14][15][16], and have attracted further interest in their role in plant health [17].
Defining the molecular markers that can assist molecular breeding or gene-editing approaches is a necessary step to accelerate breeding efforts for tree crops. WB is among the few pathosystems in which complete genome sequences of both the host and pathogen are available as a valuable reference [18][19], contributing to the molecular phenotyping of the disease process. To provide an in-depth assessment of the molecular responses and interactions between the host and pathogen leading to WB, researchers performed a comparative proteomic analysis of symptomatic vs. healthy fruit tissues. By combining state-of-the-art LC-MS/MS and data analysis methods for peptide identification, the major molecular functions and biological processes affected by WB were determined, as were the individual proteins and protein families with differential abundance. These molecular networks deepen our understanding of the role of specific proteins and protein classes in plant-pathogen interactions and reveal details of the disease mechanisms behind WB. This can potentially lead to more targeted and sustainable therapies to protect walnut production against diseases like WB.
2. Proteome Analysis of Walnut Bacterial Blight Disease
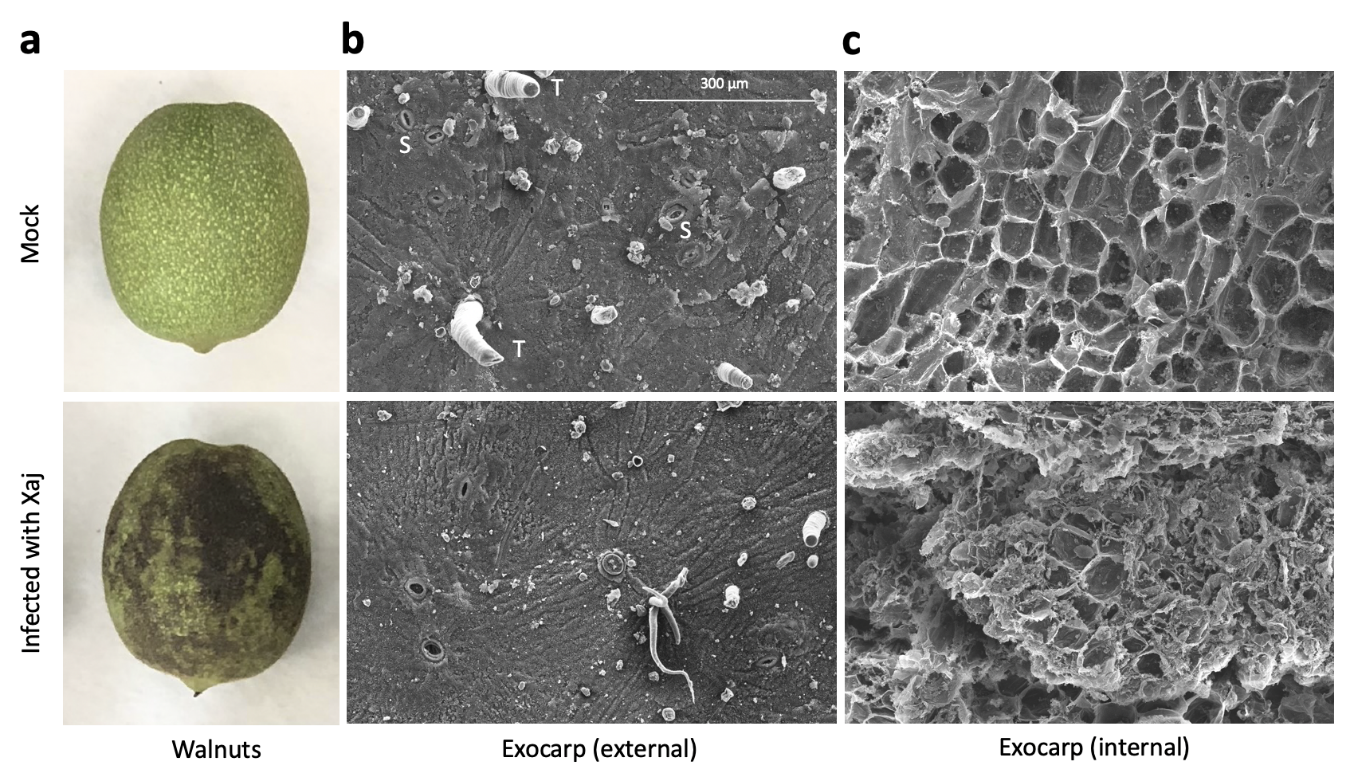
Controlled Xaj inoculations of walnut fruits closely replicated the typical WB phenotype in the disease symptom development (Figure 1a). The symptom development was quantified and compared to the mock inoculations (Figure S1), followed by microscopy of the exocarp (hull) to determine how the infection affects the surface and cell wall structures of the external-facing and internal layers below the surface of the exocarp (Figure 1b,c).
Figure 1. Walnut blight symptoms in walnut fruits. Samples were inoculated with 1 × 108 bacterial cells/mL in a solution of MgCl2 and incubated in a box to maintain high humidity at room temperature for ten days under a 12-hour light/dark cycle. (a) Walnut hull symptom development after seven days. (b) Scanning electron micrograph of the external and (c) internal-facing exocarp tissues (250×) after ten days. S: stomata and T: trichome.
2.1. Quantitative proteomic analysis of Juglans regia in response to Walnut Blight
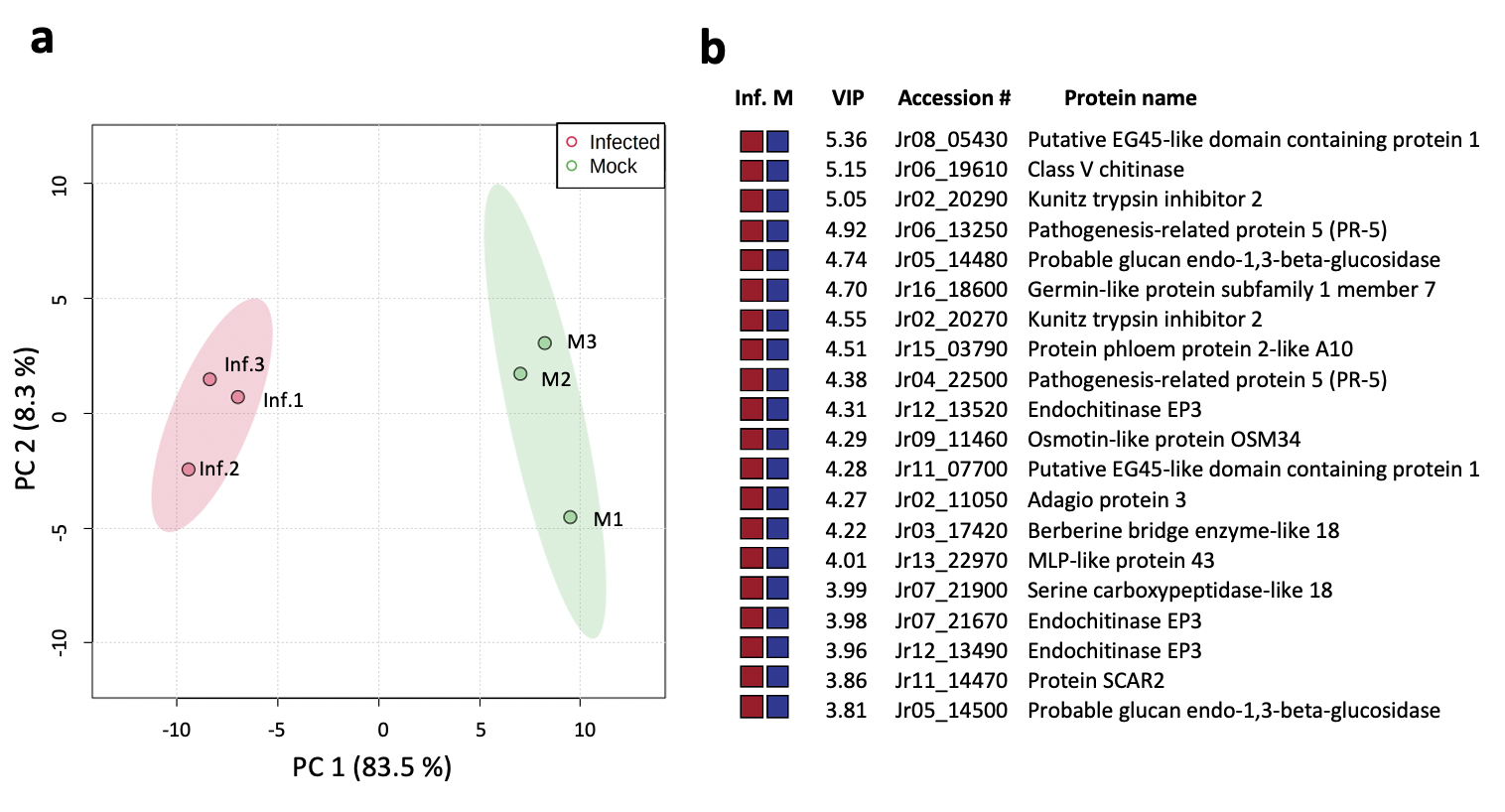
For an unbiased analysis of the proteomic results, the six samples were clustered using principal component analysis (PCA) on the 3296 identified Jr proteins. The two main components explained 91.8% (PC1 83.5% and PC2 8.3%) of the variation between the infected and mock samples, forming two distinct tight clusters according to the sample group (Figure 3a). The heatmap visualization of the proteomic data also showed distinct segregation between the infected and mock-inoculated groups (Figure S2a). The supervised partial least squares-discriminant analysis (PLS-DA) measured the variable importance in projection (VIP) score and estimated the importance of each protein in the PLS model (Figure S2b,c), increasing the relevant biomarker discovery that best explains the difference between the two sample groups. This analysis allowed the identification of the top 20 proteins responsible for group discrimination (Figure 3b).
Figure 2. Juglans regia proteome analysis comparing the infected and mock-inoculated samples. (a) Principle component analysis (PCA) plot with distinct groups and samples. (b) Top 20 proteins contributing to the variance between the observed partial least squares-discriminant analysis (PLS-DA) (Figure S2a) with high variable importance in projection (VIP) scores (Table S3), with colored boxes indicating the relative abundance (greater: red and reduced: blue) of the corresponding protein detected in infected (Inf.) and mock-inoculated (M) tissues.
2.2. Quantitative Proteomic Analysis of Xanthomonas arboricola pv. juglandis in Walnut Hulls
An analysis of WB symptomatic tissue revealed 676 Xaj proteins. Protein abundances from the three replicates representing six nuts were averaged and ranked from the most to the least abundant (Table S2). Due to the number of replicates, the proteins that were not detected in all three pools (117) were removed; some could have biological relevance, such as lipase, chitinase, hemolysin-like, and the copper homeostasis protein CutC. The 20 most abundant differential proteins were determined (Table 1). The most abundant are the OmpA-OmpF, and the list includes well-known PAMPs Ef-Tu and Ax21, GroEL, GroES, glucose dehydrogenase, tetratricopeptide repeat protein, peptidylprolyl isomerase, VirK-like, siderophore, TonB-dependent receptor, glutamine synthetase, ATP synthase, succinyl-CoA synthetase, DNA-binding protein, malate dehydrogenase, peroxiredoxin, cold-shock protein, and a hypothetical protein AKJ12_RS21505 (Table 1).
Table 1. Top 20 most abundant Xaj 417 proteins in infected walnut hulls. SP: signal peptide
2.3. Gene ontology annotation and enrichment analysis
An analysis of the 272 Jr proteins with reduced abundance in the infected tissues (log2 FC < -1 and FDR ≤ 0.05) resulted in 237 mapped proteins overrepresented in biological processes such as chromatin silencing (GO:0006342), ribosomal large subunit biogenesis (GO:0042273), and the monosaccharide metabolic process (GO:0005996), among 61 other classified terms, suggesting an arrest in the biosynthesis and channeling of the carbon and energy sources toward the degradation pathways. Eight molecular functions decreased during infection, including the structural constituents of the ribosome (GO:0003735), RNA binding (GO:0003723), and oxidoreductase activity (GO:0016491). The cellular components most negatively affected among the 29 terms included the ribosomal subunit (GO:0044391), chloroplast (GO:0009507), and cytosol (GO:0005829). The eight PANTHER protein classes highlighted included histone (PC00118), the ribosomal protein (PC00202), and dehydrogenase (PC00092). No specific pathway was statistically overrepresented in the protein set with the reduced abundance in symptomatic tissues.
3. Conclusion
Walnut bacterial blight is the most serious above-ground bacterial disease of this tree crop, as the pathogen targets nut tissues, devastating the productivity and quality of walnut orchards. This is the first study that examines the molecular responses during disease development by comparing the proteomes of infected fruit hulls to healthy tissue. This study was leveraged by the complete genome sequences from the pathogen and the recently published chromosome-scale reference walnut genome [20] to more accurately predict the contribution of the specific plant or pathogen-derived proteins and determine their role in WB disease. The degradation of plant tissues in infected fruits, shown by the scanning electron microscopic analysis, can now be interpreted from a proteomic perspective, as formed by the plant-pathogen interaction. Describing the contributions and interplay of thousands of individual proteins is no trivial task; however, the proteome data identifies major contributors that deserve further discussion and characterization.
This entry is adapted from the peer-reviewed paper 10.3390/ijms21207453
References
- Lee, Y.-A. Increased toxicity of iron-amended copper-containing bactericides to the walnut blight pathogen Xanthomonas campestris pv. juglandis. Phytopathology 1993, 83, 1460.
- Haack, S.E.; Wade, L.; Förster, H.; Adaskaveg, J.E. Epidemiology and management of bacterial spot of almond caused by Xanthomonas arboricola pv. pruni, a new disease in California. Plant Dis. 2020, 104, 1685–1693.
- Yang, B.; Sugio, A.; White, F.F. Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. USA 2006, 103, 10503–10508.
- Schornack, S.; Meyer, A.; Römer, P.; Jordan, T.; Lahaye, T. Gene-for-gene-mediated recognition of nuclear-targeted AvrBs3-like bacterial effector proteins. J. Plant Physiol. 2006, 163, 256–272.
- Tamir-Ariel, D.; Navon, N.; Burdman, S. Identification of genes in Xanthomonas campestris pv. vesicatoria induced during its interaction with tomato. J. Bacteriol. 2007, 189, 6359–6371.
- Seo, Y.-S.; Chern, M.; Bartley, L.E.; Han, M.; Jung, K.-H.; Lee, I.; Walia, H.; Richter, T.; Xu, X.; Cao, P.; et al. Towards establishment of a rice stress response interactome. PLoS Genet. 2011, 7, e1002020.
- Boulanger, A.; Zischek, C.; Lautier, M.; Jamet, S.; Rival, P.; Carrère, S.; Arlat, M.; Lauber, E. The plant pathogen Xanthomonas campestris pv. campestris exploits N-acetylglucosamine during infection. mBio 2014, 5, e01527-14.
- Sels, J.; Mathys, J.; De Coninck, B.; Cammue, B.P.A.; De Bolle, M.F. Plant pathogenesis-related (PR) proteins: A focus on PR peptides. Plant Physiol. Biochem. 2008, 46, 941–950.
- Zipfel, C. Plant pattern-recognition receptors. Trends Immunol. 2014, 35, 345–351.
- Cui, H.; Tsuda, K.; Parker, J.E. Effector-triggered immunity: From pathogen perception to robust defense. Annu. Rev. Plant Biol. 2015, 66, 487–511.
- Gottig, N.; Vranych, C.V.; Sgro, G.G.; Piazza, A.; Ottado, J. HrpE, the major component of the Xanthomonas type three protein secretion pilus, elicits plant immunity responses. Sci. Rep. 2018, 8, 9842.
- Jiang, S.; Balan, B.; Assis, R.A.B.; Sagawa, C.H.D.; Wan, X.; Han, S.; Wang, L.; Zhang, L.; Zaini, P.A.; Walawage, S.L.; et al. Genome-wide profiling and phylogenetic analysis of the SWEET sugar transporter gene family in walnut and their lack of responsiveness to Xanthomonas arboricola pv. juglandis infection. Int. J. Mol. Sci. 2020, 21, 1251.
- Lindow, S.; Olson, W.; Buchner, R. Colonization of dormant walnut buds by Xanthomonas arboricola pv. juglandis is predictive of subsequent disease. Phytopathology 2014, 104, 1163–1174.
- Zekiri, F.; Molitor, C.; Mauracher, S.G.; Michael, C.; Mayer, R.L.; Gerner, C.; Rompel, A. Purification and characterization of tyrosinase from walnut leaves (Juglans regia). Phytochemistry 2014, 101, 5–15.
- Martínez-García, P.J.; Crepeau, M.; Puiu, D.; Gonzalez-Ibeas, D.; Whalen, J.; Stevens, K.A.; Paul, R.; Butterfield, T.S.; Britton, M.T.; Reagan, R.L.; et al. The walnut (Juglans regia) genome sequence reveals diversity in genes coding for the biosynthesis of non-structural polyphenols. Plant J. 2016, 87, 507–532.
- Glagoleva, A.Y.; Shoeva, O.Y.; Khlestkina, E.K. Melanin pigment in plants: Current knowledge and future perspectives. Front. Plant Sci. 2020, 11, 770.
- Araji, S.; Grammer, T.A.; Gertzen, R.; Anderson, S.D.; Mikulic-Petkovsek, M.; Veberic, R.; Phu, M.L.; Solar, A.; Leslie, C.A.; Dandekar, A.M.; et al. Novel roles for the polyphenol oxidase enzyme in secondary metabolism and the regulation of cell death in walnut. Plant Physiol. 2014, 164, 1191–1203.
- Pereira, U.P.; Gouran, H.; Nascimento, R.; Adaskaveg, J.E.; Goulart, L.R.; Dandekar, A.M. Complete genome sequence of Xanthomonas arboricola pv. juglandis 417, a copper-resistant strain isolated from Juglans regia L. Genome Announc. 2015, 3, e01126-15.
- Marrano, A.; Britton, M.; Zaini, A.P.; Zimin, A.V.; Workman, E.R.; Puiu, D.; Bianco, L.; Di Pierro, E.A.; Allen, B.J.; Chakraborty, S.; et al. High-quality chromosome-scale assembly of the walnut (Juglans regia L.) reference genome. GigaScience 2020, 9, giaa050.
- Marrano, A.; Britton, M.; Zaini, A.P.; Zimin, A.V.; Workman, E.R.; Puiu, D.; Bianco, L.; Di Pierro, E.A.; Allen, B.J.; Chakraborty, S.; et al. High-quality chromosome-scale assembly of the walnut (Juglans regia L.) reference genome. GigaScience 2020, 9, giaa050.