The gut–liver-axis is a bidirectional coordination between the gut, including microbial residents, the gut microbiota, from one side and the liver on the other side. Any disturbance in this crosstalk may lead to a disease status that impacts the functionality of both the gut and the liver. A major cause of liver disorders is hepatitis C virus (HCV) infection that has been illustrated to be associated with gut microbiota dysbiosis at different stages of the disease progression. This dysbiosis may start a cycle of inflammation and metabolic disturbance that impacts the gut and liver health and contributes to the disease progression. This review discusses the latest literature addressing this interplay between the gut microbiota and the liver in HCV infection from both directions. Additionally, we highlight the contribution of gut microbiota to the metabolism of antivirals used in HCV treatment regimens and the impact of these medications on the microbiota composition.
- gut microbiota
- gut liver axis
- HCV
- Dysbiosis
- microbiome
1. Gut–Liver Axis
The gut–liver axis represents the link between the gut microbiota and an extraintestinal organ, the liver, where both communicate via the portal vein, systemic circulation, and biliary tract[1]. Portal circulation through the portal vein is responsible for most anatomical interactions and communications between the liver and the gut. It supplies about 70% of the blood to the liver and, therefore, is responsible for transporting nutrients and metabolites produced in the gut to the liver[2]. The gut microbiota and host cells metabolize nutritional macromolecules such as carbohydrates, lipids, and proteins. The metabolic products may be transferred to the liver through the portal vein[3]. This communication also leads to the transport of toxic byproducts of the gut microbiota to the liver such as peptidoglycans, endotoxins, or the intact bacteria, which may disrupt metabolic functions of the liver[4]. On the other side, bile acids are generated in the liver and then released into the intestine, and therefore they get involved in the regulation and communication of the gut–liver axis[5][6]. Portal circulation through the portal vein is responsible for most anatomical interactions and communications between the liver and the gut. It supplies about 70% of the blood to the liver and, therefore, is responsible for transporting nutrients and metabolites produced in the gut to the liver[7]. The gut microbiota and host cells metabolize nutritional macromolecules such as carbohydrates, lipids, and proteins. The metabolic products may be transferred to the liver through the portal vein[8]. As mentioned above, this communication also leads to the transport of harmful byproducts of the gut microbiota to the liver, which may disrupt metabolic functions of the liver[9]. On the other side, bile acids are generated in the liver and then released into the intestine, and therefore they get involved in the regulation and communication of the gut–liver axis [10].
1.1. Bile Acids and Gut-Liver Axis
Synthesis of bile acids occurs in the liver from cholesterol to produce the primary bile acids, including cholic acid and chenodeoxycholic acid. These bile acids are transported to the duodenum by the biliary tract where they are converted by 7α-dehydroxylation and deconjugation into secondary bile acids including deoxycholic acid and lithocholic acid. Such bile acid conversion is mediated by some gut microbes, mainly Clostridiales[11][12]. Bile acids have crucial roles in food digestion, keeping the integrity of gut mucosa and having antimicrobial activity against invading pathogens[12][13]. It is well reported that late complicated cases of HCV infections, especially cirrhosis, are characterized by alteration of the conversion of primary bile acids into secondary bile acids; however, the mechanism of this alteration is still unknown[14]. One scenario that may explain the bile acid reduction with the progression of HCV infection may be because of gut microbiota dysbiosis, which eventually leads to decreasing the microbial diversity and increasing the abundance certain microbial taxa such as the Proteobacteria phylum, Enterobacteriaceae family, and the genera Staphylococcus and Enterococcus[14] [15].
Bile acids have attracted considerable research interest in HCV treatment due the reported defect in the treatment with IFN therapies in patients who have high serum levels of bile acids[16][17]. On the other side, HCV treatment with direct-acting antiviral (DAA) was associated with a four-fold increase in the total bile acid levels by week 4 antiviral therapy. Moreover, ritonavir was reported to induce changes in bile acid transport. Regarding the functions of bile acids, it is well known that bile salts play a vital role in absorption and digestion of lipid components in addition to their role as ligands for nuclear receptor known as farnesoid X receptor (FXR) that controls several genes required for the metabolism of different macromolecules such as lipids, glucose, and bile acids[18] . It was reported that bile salts lead to activation of FXR that was associated with the activation of lipoprotein lipase, which induces entry of HCV to different cell types and interferes with its infection[19]. Another study reported that bile acids induce HCV replication depending on EGFR/ERK pathway that affects the efficacy of anti-viral treatment drugs[20]. There are previous studies that reported the role of bile acids in the replication of HCV genotype 1[21][22]. Another study detected the ability of bile acids to increase genotype 1 and genotype 2a replication in Huh7 and Huh6 cell lines, but with lower capacity in the case of genotype 2a that suggest the interference of bile salts with anti-viral treatment and their role to improve HCV replication in vitro in cell lines [23].
1.2. Intestinal Barrier
The intestinal barrier prevents toxic compounds such as bacteria and their byproducts from reaching extraintestinal body organs and other tissues including the liver. Thus, impaired intestinal barrier exposes the liver to harmful and toxic compounds from the intestine, which may cause liver damage and impairment of its function, such as alcoholic liver disease, primary biliary cholangitis, and liver cirrhosis [24]. For instance, the increase in intestinal permeability, associated with gut microbiota dysbiosis, exposes hepatocytes pathogen-associated molecular patterns (PAMP), and damage-associated molecular patterns that contribute to liver injury[25]. PAMPs have a direct effect either on hepatocytes or innate immune cells in the liver, such as kupffer and stellate cells [26]. Accordingly, the above discussed findings illustrate that the relationship between the liver and the gut microbiota is bidirectional, and gut microbiota dysbiosis is associated with liver disorders. However, the mechanistic understanding of the cause–effect relationship requires further studies on a case-by-case fashion.
2. Gut Microbiota Dysbiosis in HCV
2.1. Gut Microbiota During Liver Disease Manifestations
Gut microbiota have been linked to fatty liver disease, autoimmune hepatitis, alcoholic liver disease, viral hepatitis (HBV and HCV), primary biliary cholangitis, and primary sclerosing cholangitis (PSC) [26][27][28][29][30][31]. Some gut microbes such as Lactobacillus plantarum, Saccharomyces boulardii, and Bifidobacterium species may mitigate different types of hepatitis and metabolic disorders [32]. The gut mucous membranes act as the entrance doors for invading pathogens[33]. In the case of viruses causing hepatitis, the virus shatters the intestinal mucosa and breaches the permeability, resulting in gut dysbiosis and proclamation of liver cirrhosis and HCC through inducing the release of pro-inflammatory cytokines[34]. Therefore, it is not surprising that mitigation of gut dysbiosis, for example, by using probiotics or prebiotics, helps lessen the tolerogenic response and improve mucosal protection against viral pathogens[35]. For instance, the increased abundance of Lactobacillus in the gut microbiota positively correlates with murine norovirus inhibition by vitamin A, through the upregulation of interferon (IFN-β) [36]. Moreover, a mixture of Bifidobacterium and various probiotics with fructo-oligosaccharides and galacto-oligosaccharides has a protective effect against Rotavirus infection in a rat model [35]. This is mediated by upregulating the expression of IFN-γ, IL-4, TNF-α, and TLR2 promoting their production[35]. The major complications of liver diseases, notably cirrhosis, are characterized by gut dysbiosis depicting an increase in the families of Enterobacteriaceae, Veillonellaceae, and Proteobacteria phylum, as well as a decrease in Lachnospiraceae family and the phylum of Bacteroidetes [37]. Consistently, the cirrhosis dysbiosis ratio is derived to describe the variations in gut microbiota in patients suffering from cirrhosis with beneficial Ruminococcaceae and Lachnospiraceae and harmful Enterobacteriaceae bacteria [38]. Other liver manifestations involving severe cirrhosis and hepatic encephalopathy are characterized by an elevation in Enterobacteriaceae levels[39].
2.2. Dysbiosis of Gut Microbiota During Hepatic Viral Manifestations
Hepatitis occurs due to infection with different viruses, including hepatitis A (HAV), HBV, HCV, or hepatitis E virus (HEV). Hepatitis represents an important health concern, particularly in developing countries[34]. HAV and HEV lead to acute manifestations that may be self-cured and short-lived except in immunosuppressed individuals. Taxonomically, HAV and HEV belong to RNA viruses that can be transmitted via ingestion of contaminated food and water and may severely impact intestinal microbiota[40]. A previous study demonstrated that HEV’s manifestations in pigs can be treated using probiotics containing Enterococcus faecium NCIMB 10415[41]. Nonetheless, there is a deficiency of related records in humans. Besides, HAV and HEV, HBV, and HCV are serious health problems globally that are responsible for numerous reported cases of chronic hepatitis and death[42][43]. The major concern regarding these two viruses is their ability to establish chronic infections (at 10% in HBV and more than 30% in HCV) that finally result in severe complications, including cirrhosis and HCC[42][43]. In order to evade the immune system, hepatic viruses have developed many mechanisms, including dysbiosis of intestinal microbiota [44]. It is well documented that chronic hepatitis infections lead to massive translocation of the intestinal microbiota [45][46]. Such translocation impairs the primary barrier causing the growth of pathogenic bacteria at high rates, and abnormal regulation of immune cells that finally results in severe intestinal inflammation [47]. Moreover, the situation may worsen due to loss of homeostasis of the intestinal microbiota, which results in the progression of hepatitis viral infection [48]. Therefore, during chronic hepatic viral infections, microbiota have a great influence on viral replication as well as the interactions between host cells and the virus. During viral hepatitis, some taxa prevail including the family of Enterobacteriaceae and the species including Enterococcus faecalis, Escherichia coli, and Faecalibacterium prausnitzii, while others are reduced, such as lactic acid bacteria involving the genera of Leuconostoc, Lactobacillus, Weissella, and Pediococcus[38][49]. Moreover, prevalence of other bacterial species are related to liver complications (cirrhosis and/or HCC) following HBV or HCV infections, including bacterial families such as Enterobacteriaceae, genera including Neisseria and Gemella, and species like E. faecalis, E. coli, and F. prausnitzii [42][43]. Finally, a fungal pathogen, namely Candida, is reported in patients suffering from HBV related cirrhosis [50].
2.3. Dysbiosis of Gut Microbiota During HCV Infection
HCV is considered one of the major etiological agents of hepatitis, which results in severe liver complications, including cirrhosis, HCC, and it may even lead to liver failure and death[41]. There is little published literature regarding the impact of HCV infection on gut microbiota[9][37][51][52][53][54][55][56]. Additionally, the mechanistic understanding of how microbial dysbiosis participates in the disease’s progression is not completely addressed. It is well documented that dysbiosis can be classified into three main categories; (i) alteration of beneficial microorganisms, (ii) predominance of harmful microorganisms or pathobionts, and (iii) alteration of total microbial diversity[57]. The studies performed to reveal the effect of HCV on microbiota dysbiosis depicted lower bacterial diversity in HCV infected patients than healthy individuals. This diversity alteration is directly related to the severity and stage of the disease[28]. The previous finding may be attributed to immunoglobulin A (IgA) production by gastric B-lymphocytes infected by HCV that induces changes in the constitution of gut microbiota [58]. Interestingly, chronic liver diseases in advanced stages are associated with more alterations in gut microbiota than patients with the less developed disease[59]. This HCV-associated depletion of the gut microbiota diversity was mitigated after treatment with antiviral drugs[15]. In contrast, a recent study reported a lower microbial diversity in healthy adults as compared to treatment of naïve newly diagnosed HCV patients, which illustrates the impact of the treatment as a confounding factor in microbiome studies[54]. Additionally, this difference in HCV-associated diversity may be attributed to different cohort’s ethnicities, therapeutic factors, and different disease stages.
It was observed that during HCV infection, the order Clostridiales is significantly depleted, while both Lactobacillus and Streptococcus genera are significantly increased[34]. In addition, it is well documented that the phylum of Bacterioidetes, the family of Enterobacteriaceae, and viridans streptococci are increased in the case of chronic HCV patients, while the phylum of Firmicutes is decreased[28]. It is also noteworthy that HCV is associated with an increase in the lipopolysaccharides (LPS) serum levels, which indicates a damaged intestinal barrier and microbial translocation and inflammation during the disease progression[28][54].
The stages of HCV infection can be classified into (i) persistently normal serum alanine aminotransferase stage (PNALT), (ii) chronic hepatitis, (iii) liver cirrhosis, and (iv) HCC [28]. It is reported that there is a significant reduction in Ruminococcaceae and Lachnospiraceae families in chronic hepatitis, liver cirrhosis, and HCC patients. On the other hand, there is a significant increase in Lactobacilli and viridans streptococci. PNALT patients depict a significant increase in the genus of Bacteroides and the family of Enterobacteriaceae. Remarkably, Streptococcus salivarius is significantly increased in HCC patients with liver cirrhosis, indicating that S. salivarius may play a role in developing liver cirrhosis and progression to HCC[27]. Metabolic dysfunction of bile acids associated with HCV may be the key factor in the gut microbiota dysbiosis. This bile disturbance may result in overgrowth of proinflammatory bacteria, involving Porphyromonadaceae and Enterobacteriaceae families along with reducing the phylum of Firmicutes, the key producers of secondary bile acid generation[55]. In addition, Ruminococcaceae and Lachnospiraceae, the two Firmicutes families, are major SCFAs producers through fermentation of carbohydrates in human intestines[60][61]. Such SCFAs are crucial for the induction and differentiation of colonic regulatory T (Treg) cells responsible for suppressing inflammation [62][63]. SCFAs are also crucial for the nutrition of colon epithelia and adjusting its pH[64]. Thus, deprivation of SCFAs as a result of gut dysbiosis due to alteration of Ruminococcaceae and Lachnospiraceae would result in worsening the state of chronic HCV patients. This is mainly due to the loss of pH control, leading to hyperammonemia and ammonia absorption in the gut[65].
During both PNALT and liver cirrhosis, there is a significant increase in the genus of Bacteroides and Enterobacteriaceae family[15][43][66]. These reports explain that gut dysbiosis results in decreasing bile acids that finally alter the gut microbiota diversity [67]. In addition, the increase in Bacteroides and Enterobacteriaceae may explain the inflammation that occurs in patients suffering from hepatic encephalopathy[62][68]. Therefore, the increase in levels of Enterobacteriaceae as well as Bacteroides, may be used as a biomarker for proinflammation that finally may result in endotoxemia.
S. salivarius is significantly enriched during liver cirrhosis and HCC, suggesting that S. salivarius plays a pivotal role in the progression of chronic hepatitis into liver cirrhosis and even the development of HCC [55]. S. salivarius has down-regulatory impacts on innate immune responses; therefore, their presence may speed up the progression of HCC[66]. Table 1 summarizes the gut microbiota dysbiosis in different stages of HCV infection observed in clinical studies. Moreover, major dysbiosis of gut microbiota during HCV infection and its impact on worsening the disease’s condition and progression to cirrhosis and HCC is summarized in Figure 1.
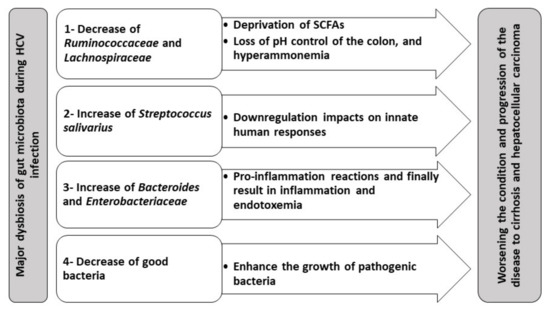
Figure 1. Gut microbiota dysbiosis in HCV and its hypothetical contribution to the progression of gut–liver axis complications.
Therefore, gut dysbiosis is directly linked to the progression of chronic HCV, probably via endotoxemia and hyperammonemia. Thus, gut dysbiosis can be a novel therapeutic target for alleviating the complications of chronic HCV. This can be achieved by either using fecal microbiota transplantation (FMT) or prebiotics and probiotics. Thus, it is not surprising that many reports have recommended the utilization of FMT with treatment regimens of fatty liver disease, PSC, liver cirrhosis, and HCC[69][70][71]. Other studies suggested the incorporation of a broad-spectrum antimicrobial as rifaximin to reduce endotoxemia and harmful metabolites [72][73], as well as to compensate for the reduction in the bile acid levels[14] that all attributed to Porphyromonadaceae, Enterobacteriaceae, and Bacteroidaceae[73] Thus, there are recommendations for using rifaximin followed by FMT as an augmenting therapy of HCV, stating that this would help get rid of S. salivarius accompanied by restoring the healthy microbiota.
Moreover, it is reported that there is no direct effect of HCV treatment using ribavirin (RBV) combined with the immune modulator pegylated interferon (PEG-IFN) on gut dysbiosis[50] Mechanistically, this regimen elevates bile acid production that is essential for gut homeostasis[14]. Therefore, we can postulate that direct-acting antivirals (DAAs) may correct for the reduced bile acid levels induced by some liver cirrhosis-associated microbiota, including the family of Enterobacteriaceae, the genera including Staphylococcus and Enterococcus[14]. HCV complications treatment can also be enhanced through utilizing Bifidobacterium and lactobacillus acidophilus, which have probiotic attributes [74]. The beneficial uses of probiotics in treatment HCV-infected patients with cirrhosis are well documented [75]. Moreover, some microbiota can enhance the immune response in HCV patients through activation of CD56+ NK cell counts and CD3+ cells [53]. Enhancement of the cytotoxic activity of NK cells can inhibit HCV replication.
Along with being a novel therapeutic approach, microbiota transition could serve as a biological indicator for the determination of chronic HCV progression. For example, this can be achieved by detecting the significant increase in Bacteroides and Enterobacteriaceae during mild liver disease or the significant increase in lactobacilli and viridans streptococci are associated with the progression of the disease[28]. Sultan and coauthors have recently identified 5 microbiota OTUs that can differentiate HCV patients from healthy individuals[54].
This entry is adapted from the peer-reviewed paper 10.3390/biology10010055
References
- Tremaroli, V.; Bäckhed, F. Functional interactions between the gut microbiota and host metabolism. Nat. Cell Biol. 2012, 489, 242–249, doi:10.1038/nature11552.
- Cremer, J.; Arnoldini, M.; Hwa, T. Effect of water flow and chemical environment on microbiota growth and composition in the human colon. Proc. Natl. Acad. Sci. USA 2017, 114, 6438–6443, doi:10.1073/pnas.1619598114.
- Jiminez, J.A.; Uwiera, T.C.; Abbott, D.W.; Uwiera, R.R.E.; Inglis, G.D. Impacts of resistant starch and wheat bran consump-tion on enteric inflammation in relation to colonic bacterial community structures and short-chain fatty acid concentrations in mice. Gut Pathog. 2016, 8, 1–20, doi:10.1186/s13099-016-0149-6.
- Zheng, X.; Qiu, Y.; Zhong, W.; Baxter, S.; Su, M.; Li, Q.; Xie, G.; Ore, B.M.; Qiao, S.; Spencer, M.D.; et al. A targeted metabo-lomic protocol for short-chain fatty acids and branched-chain amino acids. Metabolomics 2013, 9, 818–827, doi:10.1007/s11306-013-0500-6.
- Jia, W.; Gao, Z.; Zhang, J.; Ye, X.; Xu, A.; Ye, J.; Jia, W. Sodium Butyrate Stimulates Expression of Fibroblast Growth Factor 21 in Liver by Inhibition of Histone Deacetylase 3. Diabetes 2012, 61, 797–806, doi:10.2337/db11-0846.
- Brandl, K.; Kumar, V.; Eckmann, L. Gut-liver axis at the frontier of host-microbial interactions. Am. J. Physiol. Liver Physiol. 2017, 312, G413–G419, doi:10.1152/ajpgi.00361.2016.
- Quesada-Vázquez, S.; Aragonès, G.; Del Bas, J.M.; Escoté, X. Diet, Gut Microbiota and Non-Alcoholic Fatty Liver Disease: Three Parts of the Same Axis. Cells 2020, 9, 176, doi:10.3390/cells9010176.
- Wang, S.-Z.; Yu, Y.; Adeli, K. Role of Gut Microbiota in Neuroendocrine Regulation of Carbohydrate and Lipid Metabolism via the Microbiota-Gut-Brain-Liver Axis. Microorganisms 2020, 8, 527, doi:10.3390/microorganisms8040527.
- Adams, D.H.; Eksteen, B.; Curbishley, S.M. Immunology of the gut and liver: A love/hate relationship. Gut 2008, 57, 838–848, doi:10.1136/gut.2007.122168.
- Chiang, J.Y. Regulation of bile acid synthesis: Pathways, nuclear receptors, and mechanisms. J. Hepatol. 2004, 40, 539–551, doi:10.1016/j.jhep.2003.11.006.
- Ridlon, J.M.; Kang, D.-J.; Hylemon, P.B. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 2005, 47, 241–259, doi:10.1194/jlr.r500013-jlr200.
- Vlahcevic; R.Z.; Buhac, I.; Bell, C.C., Jr.; Swell, L. Abnormal metabolism of secondary bile acids in patients with cirrhosis. Gut 1970, 11, 420–422.
- Swann, J.R.; Want, E.J.; Geier, F.M.; Spagou, K.; Wilson, I.D.; Sidaway, J.E.; Nicholson, J.K.; Holmes, E. Systemic gut microbi-al modulation of bile acid metabolism in host tissue compartments. Proc. Natl. Acad. Sci. USA 2010, 108 (Suppl. 1), 4523–4530, doi:10.1073/pnas.1006734107.
- Kakiyama, G.; Pandak, W.M.; Gillevet, P.M.; Hylemon, P.B.; Heuman, D.M.; Daita, K.; Takei, H.; Muto, A.; Nittono, H.; Rid-lon, J.M.; et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J. Hepatol. 2013, 58, 949–955, doi:10.1016/j.jhep.2013.01.003.
- Ponziani, F.R.; Putignani, L.; Sterbini, F.P.; Petito, V.; Picca, A.; Del Chierico, F.; Reddel, S.; Calvani, R.; Marzetti, E.; San-guinetti, M.; et al. Influence of hepatitis C virus eradication with direct-acting antivirals on the gut microbiota in patients with cirrhosis. Aliment. Pharmacol. Ther. 2018, 48, 1301–1311, doi:10.1111/apt.15004.
- Iwata, R.; Stieger, B.; Mertens, J.; Muller, T.; Baur, K.; Frei, P.; Braun, J.; Vergopoulos, A.; Martin, I.V.; Schmitt, J.; et al. The role of bile acid retention and a common polymorphism in the ABCB11 gene as host factors affecting antiviral treatment response in chronic hepatitis C. J. Viral Hepat. 2010, 18, 768–778, doi:10.1111/j.1365-2893.2010.01363.x.
- Jorquera, F.; Monte, M.J.; Guerra, J.; Sanchez-Campos, S.; Merayo, J.A.; Olcóz, J.L.; González-Gallego, J.; Marin, J.J. Useful-ness of combined measurement of serum bile acids and ferritin as additional prognostic markers to predict failure to reach sustained response to antiviral treatment in chronic hepatitis C. J. Gastroenterol. Hepatol. 2005, 20, 547–554.
- Stauber, E.R.; Fauler, G.; Rainer, F.; Leber, B.; Posch, A.; Streit, A.; Spindelboeck, W.; Stadlbauer, V.; Kessler, H.H.; Mangge, H. Anti-HCV treatment with ombitasvir/paritaprevir/ritonavir +/- dasabuvir is associated with increased bile acid levels and pruritus. Wien. Klin. Wochenschr. 2017, 129, 848–851.
- Makishima, M.; Okamoto, A.Y.; Repa, J.J.; Tu, H.; Learned, R.M.; Luk, A.; Hull, M.V.; Lustig, K.D.; Mangelsdorf, D.J.; Shan, B. Identification of a Nuclear Receptor for Bile Acids. Science 1999, 284, 1362–1365, doi:10.1126/science.284.5418.1362.
- Andréo, U.; Maillard, P.; Kalinina, O.; Walic, M.; Meurs, E.; Martinot, M.; Marcellin, P.; Budkowska, A. Lipoprotein lipase mediates hepatitis C virus (HCV) cell entry and inhibits HCV infection. Cell. Microbiol. 2007, 9, 2445–2456, doi:10.1111/j.1462-5822.2007.00972.x.
- Patton, J.B.; George, D.; Chang, K.-O. Bile Acids Promote HCV Replication through the EGFR/ERK Pathway in Repli-con-Harboring Cells. Intervirology 2011, 54, 339–348, doi:10.1159/000321452.
- Chang, K.-O.; George, D.W. Bile Acids Promote the Expression of Hepatitis C Virus in Replicon-Harboring Cells. J. Virol. 2007, 81, 9633–9640, doi:10.1128/jvi.00795-07.
- Scholtès, C.; Diaz, O.; Icard, V.; Kaul, A.; Bartenschlager, R.; Lotteau, V.; André, P. Enhancement of genotype 1 hepatitis C virus replication by bile acids through FXR. J. Hepatol. 2008, 48, 192–199, doi:10.1016/j.jhep.2007.09.015.
- Chhatwal, P.; Bankwitz, D.; Gentzsch, J.; Frentzen, A.; Schult, P.; Lohmann, V.; Pietschmann, T. Bile Acids Specifically In-crease Hepatitis C Virus RNA-Replication. PLoS ONE 2012, 7, e36029, doi:10.1371/journal.pone.0036029.
- Konturek, C.P.; Harsch, I.A.; Konturek, K.; Schink, M.; Zopf, Y. Gut-liver axis: How intestinal bacteria affect the liver. MMW Fortschr Med. 2018. 160 (Suppl. 5), 11–15.
- Konturek, C.P.; Harsch, I.A.; Konturek, K.; Schink, M.; Konturek, T.; Neurath, M.F.; Zopf, Y. Gut⁻Liver Axis: How Do Gut Bacteria Influence the Liver? Med. Sci. 2018, 6, 79.
- Chen, W.; Wei, Y.; Xiong, A.; Li, Y.; Guan, H.; Wang, Q.; Miao, Q.; Bian, Z.; Xiao, X.; Lian, M.; et al. Comprehensive Analysis of Serum and Fecal Bile Acid Profiles and Interaction with Gut Microbiota in Primary Biliary Cholangitis. Clin. Rev. Allergy Immunol. 2019, 58, 25–38, doi:10.1007/s12016-019-08731-2.
- Granito, A.; Muratori, P.; Muratori, L. Editorial: Gut microbiota profile in patients with autoimmune hepatitis-a clue for adjunctive probiotic therapy? Aliment. Pharmacol. Ther. 2020, 52, 392–394, doi:10.1111/apt.15795.
- Inoue, T.; Nakayama, J.; Moriya, K.; Kawaratani, H.; Momoda, R.; Ito, K.; Iio, E.; Nojiri, S.; Fujiwara, K.; Yoneda, M.; et al. Gut Dysbiosis Associated With Hepatitis C Virus Infection. Clin. Infect. Dis. 2018, 67, 869–877, doi:10.1093/cid/ciy205.
- Lemoinne, S.; Sabino, J.; Sokol, H. Gut microbiota in PSC: From association to possible causality. Commentary to “Gut pathobionts underlie intestinal barrier dysfunction and liver T helper 17 cell immune response in primary sclerosing chol-angitis” by Nakamoto et al., Nature Microbiology, January 2019. Clin. Res. Hepatol. Gastroenterol. 2020, 44, 123–125.
- Mohamadkhani, A. On the potential role of intestinal microbial community in hepatocarcinogenesis in chronic hepatitis B. Cancer Med. 2018, 7, 3095–3100, doi:10.1002/cam4.1550.
- Nakano, H.; Wu, S.; Sakao, K.; Hara, T.; He, J.; Garcia, S.; Shetty, K.; Hou, D.-X. Bilberry Anthocyanins Ameliorate NAFLD by Improving Dyslipidemia and Gut Microbiome Dysbiosis. Nutrients 2020, 12, 3252, doi:10.3390/nu12113252.
- Sun, S.; Wang, K.; Sun, L.; Cheng, B.; Qiao, S.; Dai, H.; Shi, W.; Ma, J.; Liu, H. Therapeutic manipulation of gut microbiota by polysaccharides of Wolfiporia cocos reveals the contribution of the gut fungi-induced PGE2 to alcoholic hepatic steatosis. Gut Microbes 2020, 12, 1830693, doi:10.1080/19490976.2020.1830693.
- Sehgal, R.; Bedi, O.; Trehanpati, N. Role of Microbiota in Pathogenesis and Management of Viral Hepatitis. Front. Cell. Infect. Microbiol. 2020, 10, 341, doi:10.3389/fcimb.2020.00341.
- Karst, S.M. The influence of commensal bacteria on infection with enteric viruses. Nat. Rev. Genet. 2016, 14, 197–204, doi:10.1038/nrmicro.2015.25.
- Rigo-Adrover, M.D.M.; Van Limpt, K.; Knipping, K.; Garssen, J.; Knol, J.; Costabile, A.; Franch, Àngels; Castell, M.; Pé-rez-Cano, F.J. Preventive Effect of a Synbiotic Combination of Galacto- and Fructooligosaccharides Mixture with Bifidobac-terium breve M-16V in a Model of Multiple Rotavirus Infections. Front. Immunol. 2018, 9, 1318, doi:10.3389/fimmu.2018.01318.
- Lee, H.; Ko, G. Antiviral effect of vitamin A on norovirus infection via modulation of the gut microbiome. Sci. Rep. 2016, 6, 25835, doi:10.1038/srep25835.
- Zamparelli, M.S.; Rocco, A.; Compare, D.; Nardone, G. The gut microbiota: A new potential driving force in liver cirrhosis and hepatocellular carcinoma. United Eur. Gastroenterol. J. 2017, 5, 944–953, doi:10.1177/2050640617705576.
- Bajaj, J.S.; Heuman, D.M.; Hylemon, P.B.; Sanyal, A.J.; White, M.B.; Monteith, P.; Noble, N.A.; Unser, A.B.; Daita, K.; Fisher, A.R.; et al. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J. Hepatol. 2014, 60, 940–947, doi:10.1016/j.jhep.2013.12.019.
- Chen, Y.; Yang, F.; Lu, H.; Wang, B.; Chen, Y.; Lei, D.; Wang, Y.; Zhu, B.; Li, L. Characterization of fecal microbial communi-ties in patients with liver cirrhosis. Hepatol. 2011, 54, 562–572, doi:10.1002/hep.24423.
- Lemon, S.M.; Ott, J.J.; Van Damme, P.; Shouval, D. Type A viral hepatitis: A summary and update on the molecular virolo-gy, epidemiology, pathogenesis and prevention. J. Hepatol. 2018, 68, 167–184, doi:10.1016/j.jhep.2017.08.034.
- Kreuzer, S.; Machnowska, P.; Aßmus, J.; Sieber, M.; Pieper, R.; Schmidt, M.F.; Brockmann, G.A.; Scharek-Tedin, L.; Johne, R. Feeding of the probiotic bacterium Enterococcus faecium NCIMB 10415 differentially affects shedding of enteric viruses in pigs. Vet. Res. 2012, 43, 58, doi:10.1186/1297-9716-43-58.
- El-Mesery, M.; El-Mowafy, M.; Elgaml, A.; Youssef, L.F.; Abed, S.Y. Correlation of Serum Soluble Fibrinogen-Like Protein 2 with Soluble FAS Ligand and Interferon Gamma in Egyptian Hepatitis C Virus-Infected Patients and Hepatocellular Carci-noma Patients. J. Interf. Cytokine Res. 2017, 37, 342–347, doi:10.1089/jir.2016.0128.
- El-Mowafy, M.; Elgaml, A.; El-Mesery, M.; Elegezy, M. Molecular analysis of Hepatitis B virus sub-genotypes and incidence of preS1/preS2 region mutations in HBV-infected Egyptian patients from Mansoura. J. Med. Virol. 2017, 89, 1559–1566, doi:10.1002/jmv.24828.
- Visvanathan, K.; Skinner, N.A.; Thompson, A.J.; Riordan, S.M.; Sozzi, V.; Edwards, R.; Rodgers, S.; Kurtovic, J.; Chang, J.; Lewin, S.; et al. Regulation of Toll-like receptor-2 expression in chronic hepatitis B by the precore protein. Hepatology 2007, 45, 102–110, doi:10.1002/hep.21482.
- Li, D.K.; Yan, P.; Chung, R.T.; Butt, A.A.; Abou-Samra, A.-B. Proton pump inhibitors are associated with accelerated devel-opment of cirrhosis, hepatic decompensation and hepatocellular carcinoma in noncirrhotic patients with chronic hepatitis C infection: Results from ERCHIVES. Aliment. Pharmacol. Ther. 2017, 47, 246–258, doi:10.1111/apt.14391.
- Lu, H.; Wu, Z.; Xu, W.; Yang, J.; Chen, Y.; Li, L. Intestinal Microbiota Was Assessed in Cirrhotic Patients with Hepatitis B Virus Infection. Microb. Ecol. 2011, 61, 693–703, doi:10.1007/s00248-010-9801-8.
- Hill, D.A.; Hoffmann, C.; Abt, M.C.; Du, Y.; Kobuley, D.; Kirn, T.J.; Bushman, F.; Artis, D. Metagenomic analyses reveal an-tibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeo-stasis. Mucosal. Immunol. 2009, 3, 148–158, doi:10.1038/mi.2009.132.
- Xu, D.; Huang, Y.; Wang, J. Gut microbiota modulate the immune effect against hepatitis B virus infection. Eur. J. Clin. Mi-crobiol. Infect. Dis. 2015, 34, 2139–2147, doi:10.1007/s10096-015-2464-0.
- Chen, Y.; Ji, F.; Guo, J.; Shi, D.; Fang, D.; Li, L. Dysbiosis of small intestinal microbiota in liver cirrhosis and its association with etiology. Sci. Rep. 2016, 6, 34055, doi:10.1038/srep34055.
- Cui, L.; Morris, A.; Ghedin, E. The human mycobiome in health and disease. Genome Med. 2013, 5, 63–12, doi:10.1186/gm467.
- Aly, A.M.; Adel, A.; El-Gendy, A.O.; Essam, T.; Aziz, R.K. Gut microbiome alterations in patients with stage 4 hepatitis C. Gut Pathog. 2016, 8, 1–12, doi:10.1186/s13099-016-0124-2.
- Bajaj, J.S.; Sterling, R.K.; Betrapally, N.S.; Nixon, D.E.; Fuchs, M.; Daita, K.; Heuman, D.M.; Sikaroodi, M.; Hylemon, P.B.; White, M.B.; et al. HCV eradication does not impact gut dysbiosis or systemic inflammation in cirrhotic patients. Aliment. Pharmacol. Ther. 2016, 44, 638–643, doi:10.1111/apt.13732.
- Merlini, E.; Cerrone, M.; Van Wilgenburg, B.; Swadling, L.; Cannizzo, E.S.; Monforte, A.D.; Klenerman, P.; Marchetti, G. As-sociation Between Impaired Vα7.2+CD161++CD8+ (MAIT) and Vα7.2+CD161-CD8+ T-Cell Populations and Gut Dysbiosis in Chronically HIV- and/or HCV-Infected Patients. Front. Microbiol. 2019, 10, 1972, doi:10.3389/fmicb.2019.01972.
- Sultan, S.; El-Mowafy, M.; Elgaml, A.; El-Mesery, M.; El Shabrawi, A.; Elegezy, M.; Hammami, R.; Mottawea, W. Alterations of the Treatment-Naive Gut Microbiome in Newly Diagnosed Hepatitis C Virus Infection. ACS Infect. Dis. 2020, doi:10.1021/acsinfecdis.0c00432.
- Petersen, C.; Round, J.L. Defining dysbiosis and its influence on host immunity and disease. Cell. Microbiol. 2014, 16, 1024–1033, doi:10.1111/cmi.12308.
- Wellhöner, F.; Döscher, N.; Tergast, T.L.; Vital, M.; Plumeier, I.; Kahl, S.; Potthoff, A.; Manns, M.P.; Maasoumy, B.; Wedemeyer, H.; et al. The impact of proton pump inhibitors on the intestinal microbiota in chronic hepatitis C patients. Scand. J. Gastroenterol. 2019, 54, 1033–1041, doi:10.1080/00365521.2019.1647280.
- Zheng, R.; Wang, G.; Pang, Z.; Ran, N.; Gu, Y.; Guan, X.; Yuan, Y.; Zuo, X.; Pan, H.; Zheng, J.; et al. Liver cirrhosis contributes to the disorder of gut microbiota in patients with hepatocellular carcinoma. Cancer Med. 2020, 9, 4232–4250, doi:10.1002/cam4.3045.
- Milosevic, I.; Vujovic, A.; Barac, A.; Djelic, M.; Korac, M.; Spurnic, A.R.; Gmizic, I.; Stevanovic, O.; Djordjevic, V.; Lekic, N.; et al. Gut-Liver Axis, Gut Microbiota, and Its Modulation in the Management of Liver Diseases: A Review of the Literature. Int. J. Mol. Sci. 2019, 20, 395, doi:10.3390/ijms20020395.
- Dolganiuc, A.; Norkina, O.; Kodys, K.; Catalano, D.; Bakis, G.; Marshall, C.; Mandrekar, P.; Szabo, G. Viral and Host Factors Induce Macrophage Activation and Loss of Toll-Like Receptor Tolerance in Chronic HCV Infection. Gastroenterology 2007, 133, 1627–1636, doi:10.1053/j.gastro.2007.08.003.
- Qin, N.; Yang, F.; Li, A.; Prifti, E.; Chen, Y.; Shao, L.; Guo, J.; Le Chatelier, E.; Yao, J.; Wu, L.; et al. Alterations of the human gut microbiome in liver cirrhosis. Nature 2014, 513, 59–64, doi:10.1038/nature13568.
- Duncan, S.H.; Louis, P.; Flint, H. Cultivable bacterial diversity from the human colon. Lett. Appl. Microbiol. 2007, 44, 343–350, doi:10.1111/j.1472-765x.2007.02129.x.
- Leone, V.; Gibbons, S.M.; Martinez, K.; Hutchison, A.L.; Huang, E.Y.; Cham, C.M.; Pierre, J.F.; Heneghan, A.F.; Nadimpalli, A.; Hubert, N.; et al. Effects of Diurnal Variation of Gut Microbes and High-Fat Feeding on Host Circadian Clock Function and Metabolism. Cell Host Microbe 2015, 17, 681–689, doi:10.1016/j.chom.2015.03.006.
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of colonic regulatory T cells by indigenous clostridium species. Science 2011, 331, 337–341, doi:10.1126/science.1198469.
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450, doi:10.1038/nature12721.
- Wong, J.M.W.; De Souza, R.; Kendall, C.W.C.; Emam, A.; Jenkins, D.J.A. Colonic Health: Fermentation and Short Chain Fat-ty Acids. J. Clin. Gastroenterol. 2006, 40, 235–243, doi:10.1097/00004836-200603000-00015.
- Vince, A.J.; McNeil, N.I.; Wager, J.D.; Wrong, O.M. The effect of lactulose, pectin, arabinogalactan and cellulose on the pro-duction of organic acids and metabolism of ammonia by intestinal bacteria in a faecal incubation system. Br. J. Nutr. 1990, 63, 17–26, doi:10.1079/bjn19900088.
- Tuomisto, S.; Pessi, T.; Collin, P.; Vuento, R.; Aittoniemi, J.; Karhunen, P.J. Changes in gut bacterial populations and their translocation into liver and ascites in alcoholic liver cirrhotics. BMC Gastroenterol. 2014, 14, 40, doi:10.1186/1471-230x-14-40.
- Wright, G.; Jalan, R. Ammonia and inflammation in the pathogenesis of hepatic encephalopathy: Pandora’s box? Hepatology 2007, 46, 291–294.
- Cosseau, C.; Devine, D.A.; Dullaghan, E.; Gardy, J.L.; Chikatamarla, A.; Gellatly, S.; Yu, L.L.; Pistolic, J.; Falsafi, R.; Tagg, J.; et al. The Commensal Streptococcus salivarius K12 Downregulates the Innate Immune Responses of Human Epithelial Cells and Promotes Host-Microbe Homeostasis. Infect. Immun. 2008, 76, 4163–4175, doi:10.1128/iai.00188-08.
- Anand, G.; Zarrinpar, A.; Loomba, R. Targeting Dysbiosis for the Treatment of Liver Disease. Semin. Liver Dis. 2016, 36, 37–47, doi:10.1055/s-0035-1571276.
- Williamson, K.D.; Chapman, R.W. New Therapeutic Strategies for Primary Sclerosing Cholangitis. Semin. Liver Dis. 2016, 36, 5–14, doi:10.1055/s-0035-1571274.
- Haque, R.T.; Barritt, A.S.T. Intestinal microbiota in liver disease. Best Pract. Res. Clin. Gastroenterol. 2016, 30, 133–142.
- Bajaj, J.S.; Heuman, D.M.; Sanyal, A.J.; Hylemon, P.B.; Sterling, R.K.; Stravitz, R.T.; Fuchs, M.; Ridlon, J.M.; Daita, K.; Mon-teith, P.; et al. Modulation of the Metabiome by Rifaximin in Patients with Cirrhosis and Minimal Hepatic Encephalopathy. PLoS ONE 2013, 8, e60042, doi:10.1371/journal.pone.0060042.
- Kalambokis, G.N.; Mouzaki, A.; Rodi, M.; Pappas, K.; Fotopoulos, A.; Xourgia, X.; Tsianos, E.V. Rifaximin Improves Sys-temic Hemodynamics and Renal Function in Patients With Alcohol-Related Cirrhosis and Ascites. Clin. Gastroenterol. Hepa-tol. 2012, 10, 815–818, doi:10.1016/j.cgh.2012.02.025.
- Kalambokis, G.N.; Mouzaki, A.; Rodi, M.; Pappas, K.; Fotopoulos, A.; Xourgia, X.; Tsianos, E.V. Rifaximin Improves Sys-temic Hemodynamics and Renal Function in Patients With Alcohol-Related Cirrhosis and Ascites. Clin. Gastroenterol. Hepa-tol. 2012, 10, 815–818, doi:10.1016/j.cgh.2012.02.025.
- Bajaj, J.S.; Heuman, D.M.; Sanyal, A.J.; Hylemon, P.B.; Sterling, R.K.; Stravitz, R.T.; Fuchs, M.; Ridlon, J.M.; Daita, K.; Mon-teith, P.; et al. Modulation of the Metabiome by Rifaximin in Patients with Cirrhosis and Minimal Hepatic Encephalopathy. PLoS ONE 2013, 8, e60042, doi:10.1371/journal.pone.0060042.
- Kalambokis, G.N.; Mouzaki, A.; Rodi, M.; Pappas, K.; Fotopoulos, A.; Xourgia, X.; Tsianos, E.V. Rifaximin Improves Sys-temic Hemodynamics and Renal Function in Patients With Alcohol-Related Cirrhosis and Ascites. Clin. Gastroenterol. Hepa-tol. 2012, 10, 815–818, doi:10.1016/j.cgh.2012.02.025.
