Newcastle disease viruses (NDVs), also known as avian paramyxoviruses type 1, are members of the genus Orthoavulavirus, species Avian orthoavulavirus 1 in the family Paramyxoviridae. They are negative-sense, single-stranded, non-fragmented RNA viruses, distributed worldwide, and they are able to infect wild, peridomestic and domestic avian species.
- Newcastle Disease Viruses
- Virulence
- Host Adaptation
1. Introduction
NDVs are highly adaptable viruses, capable of infecting over 200 avian species and of causing a diverse range of clinical outcomes in birds [1]. The NDV genome is composed of a single-stranded RNA molecule; it encodes for at least six proteins arranged in a 3′ to 5′ order: nucleocapsid protein (NP), phosphoprotein (P), matrix protein (M), fusion protein (F), hemagglutinin-neuraminidase (HN), and RNA polymerase [2,3].
Two accessory proteins (V and W) with a possible role in virulence have been described for NDV. These proteins are generated by RNA editing of the genomic region corresponding to the phosphoprotein gene region. Although the role of the W protein is still controversial, the role of the V protein in modulating virulence through a process that involves targeting the phospho-STAT1 degradation to block IFN-α signaling has been demonstrated [4,5,6,7,8]. In a more recent manuscript, a comprehensive description of the genetic variations and the molecular evolution of the P gene-edited and the accessory viral proteins is presented [9]. A role of the V protein in host range restriction is expected as it has been shown in vitro studies with human cells that it may play a role affecting host range. However, a study on the role of the V protein on evolution during the process of natural host adaptation or a study of the role of the different V protein variants on fitness to replicate in different hosts remains to be done and will not be reviewed here.
Although every protein of the virus is likely to interact with the host in one manner or another, none have been studied more deeply than the fusion protein [10,11,12,13,14,15]. This protein allows the virus membrane to fuse to the host cell membranes during infection. The amino acids at the fusion protein cleavage site interact with secreted host mucosal proteases, which cleave the fusion protein precursor into an active form, making it possible for the virions to attach, infect and replicate in multiple tissues and organs. This enhanced capacity of the cleaved fusion protein is responsible for increased virulence [11,16,17]. Viruses that lack phenylalanine and one or more of the necessary basic amino acids at the cleavage site are normally not able to systemically spread, and they are limited to replicating in the upper respiratory and enteric tracts, thus being considered to be viruses of low virulence, also described as lentogeneic or lNDV.
There are a large variety of pathogenicity tests to evaluate NDV virulence, and these are used to determine if NDVs are capable of producing disease in naïve chickens. These tests range from evaluation of the pathogenicity in eggs (mean death time, MDT), or in one-day-old naïve chicks (intracerebral pathogenicity index, ICPI), to an evaluation in adults (vaccinated and naïve challenge models on mature SPF birds) [2,16,18,19]. Lentogens or lNDV have ICPI ≤ of 0.7, and they generally produce no clinical signs or subclinical infections with minimal respiratory involvement, whereas those of moderate pathogenicities, such as mesogens or mNDV with ICPI values of 0.7 to 1.5, generally cause clinical signs of disease, but typically result in non-lethal outcomes in chickens. Viruses of high pathogenicity, called velogens o vNDV, have ICPI > 1.5 and generally cause serious disease and mortality among naïve chickens. There are two types of velogens (neurotropic or vnNDV and viscerotropic or vvNDV) that cannot be clearly distinguished with the ICPI tests and require intravenous or clinical disease tests on adult birds for differentiation [2,20]. For vaccinated chickens, infection with viruses of lNDV and mNDV pathotypes is asymptomatic, and infection with vNDV may occasionally cause disease with low mortality if the levels of immunity obtained during vaccination are not satisfactory. Vaccination reduces but does not eliminate viral replication of vNDV.
NDVs display large genetic diversity, which determines the existence of two distinct classes of viruses (class I and II) [21,22]. While all NDVs are capable of infecting chickens, only class II virulent viruses are normally capable of causing disease. Class I viruses are less diverse with only one genotype, while class II viruses are the most diverse, of remarkable economic impact, and will be further analyzed here from the point of view of host adaptation. Lentogenic viruses of class I and II are relatively common in waterfowl, gulls, and shorebirds and generally do not cause clinical disease in chickens [23,24,25,26,27]. Among class II viruses, virulent viruses are the most studied because they cause significant mortality in naïve animals, drop in egg production, and are repeatedly isolated in vaccinated poultry in endemic regions of the world [3,20]. For the most virulent viruses, immune responses attributed to vaccination are not sufficient to prevent mucosal viral replication, and viral shedding occurs at reduced levels [28,29,30]. Therefore, virulent viruses are still capable of disseminating in domestic birds worldwide, occasionally spilling over into wild birds. Virulent viruses of intermediate virulence are often maintained in wild birds such as those which are frequently isolated from double-crested cormorants (genotype XIX) in North America and Canada and occasionally from pelicans and gulls that are in close proximity to cormorants during outbreaks [31,32,33,34]. Other class II viruses with mesogenic and velogenic neurotropic phenotypes of genotypes VI and XXI are preferentially isolated from wild and domestic Columbiformes [35,36,37,38,39,40] and occasionally from spillover into poultry or other domestic birds. Velogenic viscerotropic (vvNDV) of class II genotypes V, VII, XII, XIII, XIV, XVII and XVIII are of more recent origin, are distributed worldwide and are most frequently isolated in poultry [21]. Neurotropic viruses (vnNDV) were most commonly isolated in poultry before the 70s and less frequently since then [41,42,43]; however, they have been maintained in cormorants in the wild. Except for old viruses of genotype V in psittacine, there have not been repeated isolations of vvNDV from wild birds; thus, the existence of natural wild bird reservoirs for vvNDV remains to be demonstrated.
2. Modulation of Virulence during Host Adaptation
The hypothesis that NDV viruses modulated virulence during host adaptation requires evidence of the occurrence of different types of evolutionary events such as (1) inter-host transmission; (2) origin of recent virulent viruses from older virulent viruses; (3) genetic changes consistent with adaptation to maintenance in different hosts; and (4) demonstration of changes in virulence-associated with these genetic changes. Modern techniques of phylogenetic analysis provide evidence for events 1, 2 and 3, and comparison among the pathogenicity and virulence of viruses adapted to different hosts provides evidence for type 4 events. Although many reports support the capacity of NDV to infect multiple hosts, a recent global phylodynamic analysis of avian paramyxovirus-1 done by J. T. Hicks using the complete fusion protein coding sequence has provided sufficient statistical evidence supporting NDVs capacity for inter-host transmission [64]. Hicks’ study (Figure 3) revealed that, in general, the ancestral history of class II viruses was structured by the host, with host types clustering together within genotypes. For instance, genotypes XII, XIII, XIV, XVII and XVIII were predominantly detected in domestic chickens with evidence for few transitions to other hosts, including Anseriformes, Psittaciformes, Columbiformes and other Galliformes birds. In contrast, genotypes I and VI were mostly detected in non-chicken hosts, such as Anseriformes and Columbiformes, respectively.
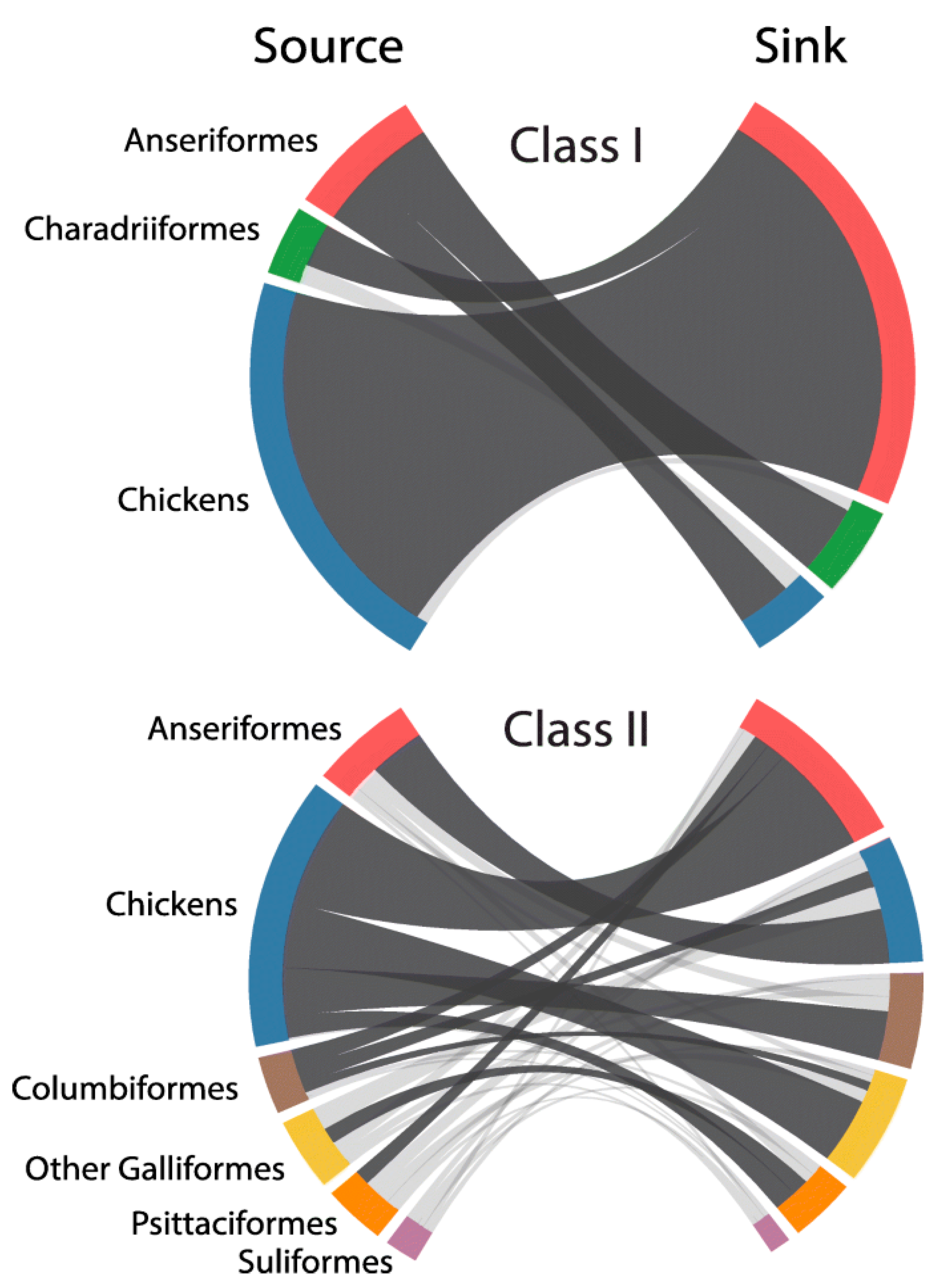
This nucleotide-based analysis demonstrated the capacity for NDV to transmit among divergent host taxa over time (Figure 3). The highest (most strongly supported) transition rate for viruses of class I and II existed from domestic chickens to Anseriformes, and among class II viruses for domestic chickens acting as a source for Columbiformes, other Galliformes, and Psittaciformes. Columbiformes was also highly supported sources of viruses to Anseriformes and domestic chickens. Notwithstanding the evidence of the existence of different transmission rates of NDV among different hosts species; some restrictions appeared to exist as virulent NDVs were not repeatedly isolated in migratory waterfowl populations (including birds of the Anatidae family ducks, swans, geese or other species with flattened beaks and fully webbed toe). Despite the abundant sampling (normally done for AIV surveillance) and of the existence of large populations of waterfowl and shorebirds in the wild, there is no strong evidence of virulent viruses being maintained in these types of birds. With only one isolated case of increased virulence for a class I virus ever reported [65], the data suggest that there may be genetic or ecological restrictions preventing the development and maintenance of highly virulent viruses in the class I viruses adapted to shorebird and waterfowl.
Demonstration of the origin of recent virulent viruses from older virulent viruses (2) and evidence of genetic changes consistent with adaptation to maintenance in different hosts (3) can be obtained by phylogenetic analysis of large databases. The genetic diversity of current NDV class II virulent genotype viruses was recently studied by an international consortium [22]. For this study, a total of 1956 complete F gene sequences were used; this included viruses of class I (n = 284) and class II (n = 1672). Detailed tables of all available sequences, including genotypes, host, date and place of isolation, are available in Supplemental Tables S1 and S2 of Dimitrov 2019 manuscript [22]. Conclusions were based on neighbor-joining, maximum-likelihood, and Bayesian method-based trees represented in their Supplementary Figure S4 for class I and Figure S5 for class II viruses [22]. Phylogenetic analysis of the fusion protein-coding region and complete genomes reaffirmed the evolution of NDVs into two large classes (class I and II). The viruses of class I were predominantly reported in wild waterfowl and shorebirds, and viruses of class II were preferentially isolated from chickens but also present in some wild bird species. Class I viruses werer lentogenic and more conserved, clustering into just one genotype, and despite the diversity in hosts, less than 10% nucleotide differences were observed among isolates. Class II viruses were between 41.0 to 46.3% genetically divergent from the class I viruses, contained the majority of virulent viruses, and had larger genetic diversity with 20 distinct genotypes and nucleotide distances among genotypes ranging from 7.8 to 28.9%.
Excerpts here are taken from Dimitrov’s full phylogenetic trees based on the complete fusion gene sequences of isolates representing NDVs of class II (n = 1672). In the phylogenetic trees produced by Dimitrov’s study, the origin of current virulent viruses can be traced back to the oldest characterized virulent genotypes circulating during the 1930s and 1940s. These include the genotype III virus identified as Australia 1932 and chicken Mukteswar 1940 as the oldest ancestors in Asia. The genotype IV oldest viruses in Europe were the Herts isolated in chickens in the United Kingdom in 1933 and the Italien isolated in Italy in 1944. In the Americas, the oldest vNDVs were members of genotype XVI and II; an example of this is the genotype XVI virus 1947 Queretaro from Mexico. In addition, the 1944 chicken California virulent virus is the oldest relative of multiple virulent viruses of genotype II isolated throughout the US. In China, the older isolates are viruses from genotype IX from 1948, forming a genotype that appears to be extinct in the wild. The phylogenetic analysis revealed that older origin genotypes, some of which are no longer circulating (e.g., genotypes II, III, IV, IX), likely may have provided indirect ancestry to present-day viruses giving rise to other genotypes such as genotype VIII.
This entry is adapted from the peer-reviewed paper 10.3390/v13010110
