Invasion and metastasis correspond to the foremost cause of cancer-related death, and the molecular networks behind these two processes are extremely complex and dependent on the intra- and extracellular conditions along with the prime of the premetastatic niche. Currently, several studies suggest an association between the levels of HOX genes expression and cancer cell invasion and metastasis, which favour the formation of novel tumour masses. The deregulation of HOX genes by HMGA2/TET1 signalling and TGFβ pathway, the HOX interference in EMT (Epithelial to Mesenchymal Transition) process and the regulatory effect of noncoding RNAs (miRs and lncRNAs) generated by the HOX loci can also promote invasion and metastasis, interfering with the expression of HOX genes or other genes relevant to these processes.
- HOX Proteins
- Metastasis
- Cancer Invasion
1. Introduction
The vast majority of cancer-related mortality in solid tumours is associated with the capacity of the cancer cells to invade and colonize nearby or distant vital organs forming metastasis [1], hallmarks that characterize the last steps of cancer progression [2]. A line of investigation suggests that tumour recurrence and metastization is led by a population of residual cells that survive treatment and are capable to leave their primary location. These cells disperse into the bloodstream, endure pressure in blood vessels, escape immune response, and acclimate to new cellular surroundings in a secondary site [3]. However, the development of new therapies that can eliminate residual tumour cells or prevent their action is conditioned by the incomplete understanding of the mechanisms underlying the long-term survival of these cells following treatment [4].
The metastatic process is complex and involves a multistep journey. The activation of invasion and metastasis is triggered by environmental stimuli, such as aging and circadian disruptions; adhesive signals from extracellular matrix (ECM) components, such as collagen and fibrin; ECM mechanical pressures, including tension and compression; cell–cell interactions; soluble signals, such as growth factors and cytokines; and intratumoural microbiota [5][6]. The invading tumour cells, on the way to the target site, interact with other proteins and cells to overcome the stromal challenges and complete the metastasis key steps of invasion, intravasation, circulation, extravasation and colonization To add more complexity to this process, secondary sites do not receive invading cancer cells passively. In fact, the host microenvironment, also designated as a premetastatic niche (PMN) or organotropic metastasis, is selectively primed by the primary tumour even before the initiation of the metastatic process. This signalling involves secretory factors and extracellular vesicles that induce vascular leakage, ECM remodelling, and immunosuppression, making the secondary microenvironment selective for the circulating tumour cells (CTCs) [7][8][9]. As common examples of organotropism, we can cite the preferred metastization of breast and prostate tumour cells to lungs, bone and liver; colon and stomach cancer to liver, lung and peritoneum; and lung carcinoma to adrenal glands and liver [10]. Hundreds of genes have been reported to lead this invasive potential, suggesting that primary tumour cells should develop a metastatic genetic signature [11]. Thus, genetic and also epigenetic modifications, accumulated during the primary tumour development and by the adaptations to the microenvironment components contribute to the metastatic process [6]. The genetic modifications involve changes in the primary DNA sequence, such as mutations, while the epigenetic mechanisms relate to chemical modifications of DNA bases and changes in the chromosomal superstructure in which DNA is packaged, such as gene promoter methylations associated with gene silencing [12][13]. In this context, alterations in HOX gene expression have been identified in primary tumours, metastasis and CTCs [14][15].
2. HOX Genes and Cancer
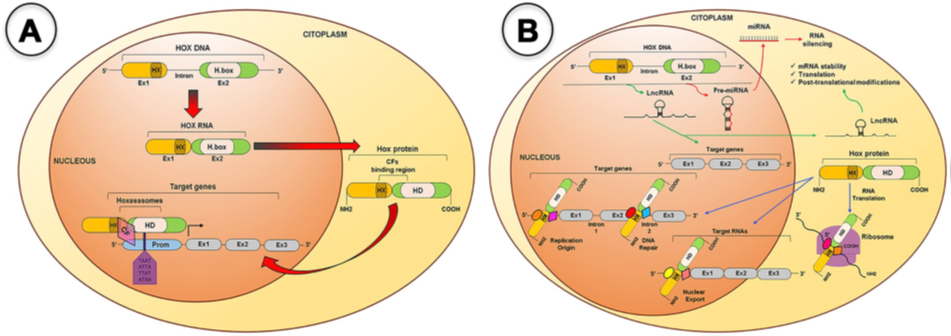
HOX genes are a family of genes that codes for transcription factors characterized by the presence of a conserved DNA sequence designated homeobox [16]. The human genome contains 39 of those genes arranged into four clusters (HOXA, HOXB, HOXC and HOXD) located in distinct chromosomes (7p15, 17q21.2, 12q13, 2q31, respectively). Each cluster presents 9–11 genes that align in 13 paralogous groups, based on sequence homology of their homeoboxes and their position within the cluster [17]. These genes are co-ordinately transcribed following temporal and spatial collinearity with an unidirectional chromatin opening, where the chromosomic order of genes is the same as the order of its expression, forming nested and overlapping expression domains [18], that define the organogenesis along the anterior–posterior axis [19][20]. HOX proteins activity occurs with the formation of multiprotein complexes, the “hoxasomes”, composed by HOX proteins, HOX cofactors and other transcription factors [15]. These cofactors are members of the three amino acids loop extension (TALE) family, which includes PBC members (PBX1–4), HMP members (MEIS1–3 and PREP1–2), and POU family (POU1–6) [21] (Figure 1). The transcription of HOX genes happens during embryonic development but also in adult cells at lower levels [22] partaking in cellular physiology and tissue homeostasis [19][23].
Figure 1. HOX proteins transcriptional and non-transcriptional roles. A. For the transcriptional roles, HOX proteins bind to promoter (Prom) regions of the target genes, alone or as a member of the Hoxasomes, in order to activate or repress target gene transcription. B. For the no transcriptional roles, several microRNAs and Long noncoding RNAs are encoded within the HOX clusters and some of them have been shown to target HOX and other genes during BC development. The HOX protein can bind to DNA or specific cofactors to modulate DNA replication and repair, mRNA nuclear export, translation and protein degradation. CF(s), Cofactor(s), Ex, Exon; H.box, Homeobox; HD, Homeodomain; HX, Hexapeptide; LncRNA, Long-noncoding RNA; mRNA, messenger RNA; miRNA, micro RNA; Prom, Promoter region [15].
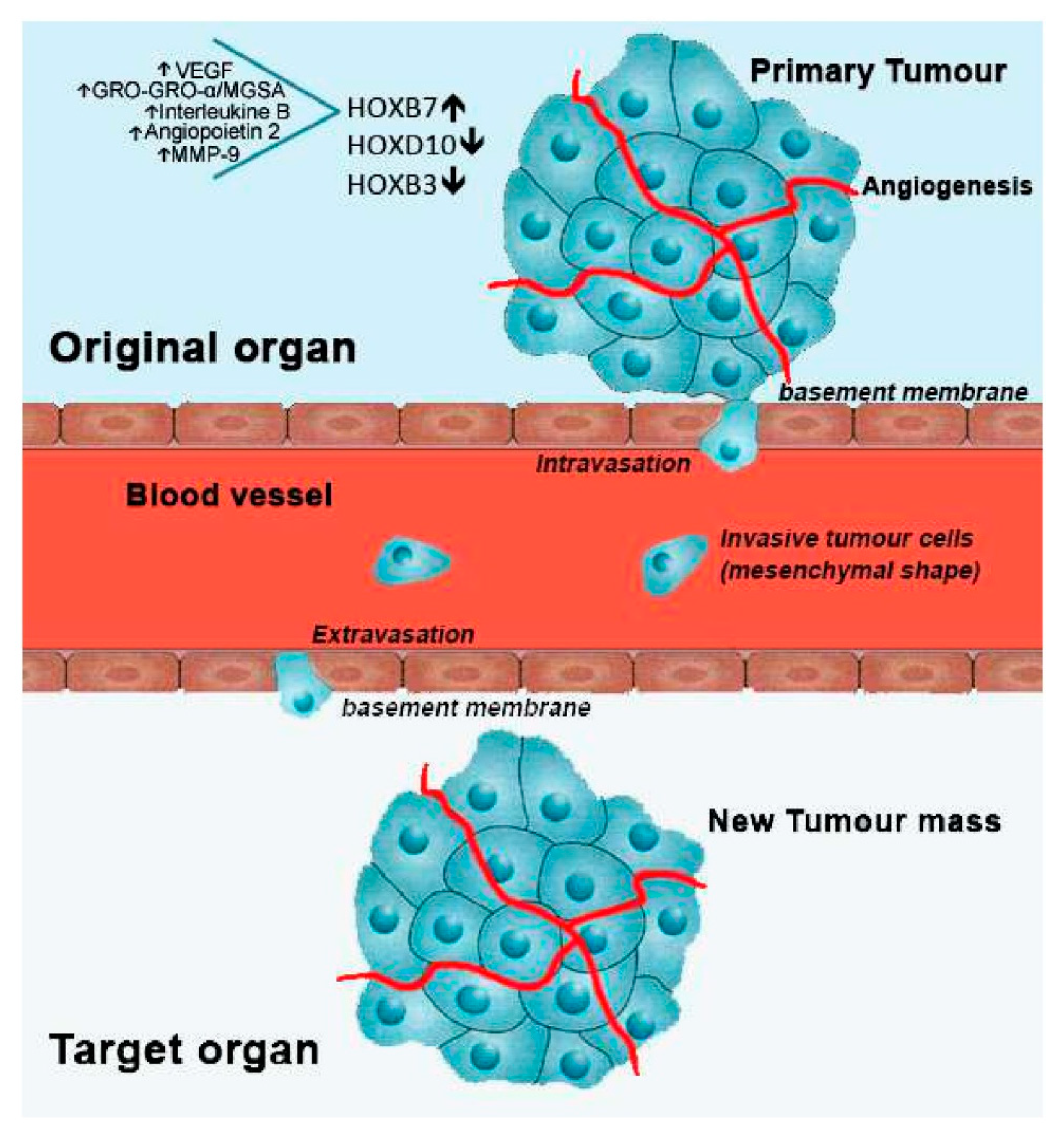
Alterations in HOX genes expression have been shown in different cancer types and associated with cancer initiation and progression [15][24][25]. They can interfere with cell differentiation, survival, proliferation, angiogenesis, autophagy, inflammation and apoptosis (Figure 2). They also influence cell movement, migration, invasion, metalloproteinase function and regulate important stemness-related genes [26][27]. More complexly, different HOX genes can activate or inhibit these processes and even exhibit antagonistic behaviours in different tissues.
Figure 2. Metastatic process and involved HOX genes, regarding breast cancer. After cell transformation, a mass is rapidly formed in a specific organ of the body, designated as primary tumour. With the formation of new blood vessels surrounding the primary tumour (angiogenesis), growth is stimulated by the flux of nutrients and oxygen. Then, cells from the primary tumour may acquire motility going through the basement membrane into the blood vessels (intravasation). Once in circulation, metastatic tumour cells stop its progress adhering to the basement membrane of a new tissue site (extravasation). Finally, cancer cells start to proliferate in the new location, forming a new tumour mass. In breast cancer, overexpression of HOXB7, along with the upregulation of the indicated molecules, and downregulation of HOXD10 and HOXB3 have been linked to the metastatic process.
Thus, HOX genes interfere with the majority of the hallmarks of cancer, whose events must occur in parallel and/or sequentially to promote cancer progression [2]. The uncontrolled cell division that represents cancer initiation, involves not only deregulated control of cell proliferation and resistance to apoptosis, but also changes in energy metabolism in order to fuel cell growth and division. For instance, normal cells under aerobic conditions process glucose, first to pyruvate via glycolysis in the cytosol and thereafter to carbon dioxide in the mitochondria; under anaerobic conditions, glycolysis is favoured. Tumour cells, even in the presence of oxygen limit their energy metabolism largely to a state of aerobic glycolysis, also known as the Warburg effect [2]. At the same time, to ensure nutrient and oxygen delivery, as well as to remove metabolic wastes and carbon dioxide from the constantly growing mass, it is fundamental to form new blood vessels through the angiogenesis process [2].
Another hallmark of cancer influenced by HOX genes, with a dual function in tumorigenesis, is inflammation [2][26]. While in normal cells, the inflammation process is carried out by innate immune cells to fight infections and heal wounds, in cancer cells it can instead inadvertently work to support tumour growth [2]. Cell movement, which drives the migration and invasion processes, is dependent on the supply of matrix-degrading enzymes such as metalloproteinases (group of enzymes that can break down proteins, such as collagen, that are normally found in the spaces between cells). Finally, the presence of cancer stem cells, with extensive growth and differentiation abilities, give phenotypic heterogeneity for tumour cells in order to enable the survival of specific cell subpopulations that are resistant to therapy and capable of regenerating the tumour once therapy has been halted [2].
An example of HOX gene alteration in cancer, relates with the increased expression of HOXA9 in the most aggressive acute leukaemia and its potential as predictive of poor prognosis [28]. Moreover, it was demonstrated that HOXA13 protein physically links to the translation initiation factor eIF4E, frequently overexpressed in cancer and described as a strong promoter of tumour growth and angiogenesis [29]. EIF4E acts on the exportation of specific oncogenes mRNAs from the nucleus to cytoplasm, such as c-Myc, FGF-2, ODC and CCND1. Thus, the deregulation of this HOX gene could facilitate the mRNA nuclear export of eIF4E-dependent oncogene transcripts, as observed in hepatocellular carcinomas (HCCs) [30]. HOX genes are also actively expressed in adipose tissue, which has emerged as an important supportive tissue for cancer proliferation and progression [26]. Half of the KRAS-mutant NSCLC (non-small-cell lung cancers) express the homeobox protein HOXC10, which triggered tumour regression in xenografts and PDX (patient-derived xenografts) models in vivo [31]. Upregulation of HOXA10 expression plays a key role in colorectal cancer development and could be considered as a new biomarker that indicates poor prognosis [32]. HOXC8 upregulation is inversely related to pancreatic ductal adenocarcinoma progression and metastasis formation and therefore could be explored as a marker for pancreatic cancer progression [33]. Moreover, some of the pathways involved in metastasis, from invasion to colonization, are affected by the HOX proteins through mechanisms that modulate their expression, such as HMGA2/TET1/HOXA and TGFβ, or by the interference of microRNAs and lncRNAs regulators [26].
Several studies are being performed to evaluate HOX proteins strength as therapeutic targets in cancer therapy and the results obtained so far have shown positive outcomes in oesophageal and oral squamous cell carcinomas [34][35], melanoma [36][37], ovarian cancer (OC) [38], breast cancer [39], meningioma [40], prostate cancer [41], and leukaemia [42]. Moreover, the use of RNA interference mechanisms [43][44] and the control of HOX methylation status [45] are additional tools that can control HOX expression with therapeutic function. However, despite the promising strategy of using HOX genes expression manipulation to prevent invasion and metastasis, further studies are required to consolidate their real potential as therapeutic targets [46].
This entry is adapted from the peer-reviewed paper 10.3390/cancers13010010
References
- Dillekås, H.; Rogers, M.S.; Straume, O. Are 90% of deaths from cancer caused by metastases? Cancer Med. 2019, 8, 5574–5576.
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674.
- Massagué, J.; Obenauf, A.C. Metastatic colonization by circulating tumour cells. Nature 2016, 529, 298–306.
- Damrauer, J.S.; Phelps, S.N.; Amuchastegui, K.; Lupo, R.; Mabe, N.W.; Walens, A.; Kroger, B.R.; Alvarez, J.V. Foxo-dependent Par-4 Upregulation Prevents Long-term Survival of Residual Cells Following PI3K-Akt Inhibition. Mol. Cancer Res. Mcr. 2018, 16, 599–609.
- Tabassum, D.P.; Polyak, K. Tumorigenesis: It takes a village. Nat. Rev. Cancer 2015, 15, 473–483.
- Fares, J.; Fares, M.Y.; Khachfe, H.H.; Salhab, H.A.; Fares, Y. Molecular principles of metastasis: A hallmark of cancer revisited. Signal Transduct. Target. Ther. 2020, 5, 28.
- Peinado, H.; Zhang, H.; Matei, I.R.; Costa-Silva, B.; Hoshino, A.; Rodrigues, G.; Psaila, B.; Kaplan, R.N.; Bromberg, J.F.; Kang, Y.; et al. Pre-metastatic niches: Organ-specific homes for metastases. Nat. Rev. Cancer 2017, 17, 302–317.
- Liu, Y.; Cao, X. Characteristics and Significance of the Pre-metastatic Niche. Cancer Cell 2016, 30, 668–681.
- Liu, Y.; Cao, X. Organotropic metastasis: Role of tumor exosomes. Cell Res. 2016, 26, 149–150.
- Meleth, S.; Whitehead, N.; Evans, T.S.; Lux, L. AHRQ Technology Assessments. In Technology Assessment on Genetic Testing or Molecular Pathology Testing of Cancers with Unknown Primary Site to Determine Origin; Agency for Healthcare Research and Quality (US): Rockville, MD, USA, 2013.
- Ramaswamy, S.; Ross, K.N.; Lander, E.S.; Golub, T.R. A molecular signature of metastasis in primary solid tumors. Nat. Genet. 2003, 33, 49–54.
- Kanwal, R.; Gupta, S. Epigenetic modifications in cancer. Clin. Genet. 2012, 81, 303–311.
- Al Aboud, N.M.; Tupper, C.; Jialal, I. Genetics, Epigenetic Mechanism. In StatPearls; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2020.
- Bhatlekar, S.; Fields, J.Z.; Boman, B.M. Role of HOX Genes in Stem Cell Differentiation and Cancer. Stem Cells Int. 2018, 2018, 3569493.
- De Bessa Garcia, S.A.; Araújo, M.; Pereira, T.; Mouta, J.; Freitas, R. HOX genes function in Breast Cancer development. Biochim. Biophys. Acta Rev. Cancer 2020, 1873, 188358.
- Scott, M.P. Vertebrate homeobox gene nomenclature. Cell 1992, 71, 551–553.
- Bhatlekar, S.; Fields, J.Z.; Boman, B.M. HOX genes and their role in the development of human cancers. J. Mol. Med. (Berl. Ger.) 2014, 92, 811–823.
- Zakany, J.; Duboule, D. The role of Hox genes during vertebrate limb development. Curr. Opin. Genet. Dev. 2007, 17, 359–366.
- Shah, N.; Sukumar, S. The Hox genes and their roles in oncogenesis. Nat. Rev. Cancer 2010, 10, 361–371.
- Luo, Z.; Rhie, S.K.; Farnham, P.J. The Enigmatic HOX Genes: Can We Crack Their Code? Cancers 2019, 11, 323.
- Longobardi, E.; Penkov, D.; Mateos, D.; De Florian, G.; Torres, M.; Blasi, F. Biochemistry of the tale transcription factors PREP, MEIS, and PBX in vertebrates. Dev. Dyn. 2014, 243, 59–75.
- Rux, D.R.; Wellik, D.M. Hox genes in the adult skeleton: Novel functions beyond embryonic development. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2017, 246, 310–317.
- Paço, A.; de Bessa Garcia, S.A.; Freitas, R. Methylation in HOX Clusters and Its Applications in Cancer Therapy. Cells 2020, 9, 1613.
- Nagata, H.; Kozaki, K.I.; Muramatsu, T.; Hiramoto, H.; Tanimoto, K.; Fujiwara, N.; Imoto, S.; Ichikawa, D.; Otsuji, E.; Miyano, S.; et al. Genome-wide screening of DNA methylation associated with lymph node metastasis in esophageal squamous cell carcinoma. Oncotarget 2017, 8, 37740–37750.
- Sui, B.Q.; Zhang, C.D.; Liu, J.C.; Wang, L.; Dai, D.Q. HOXB13 expression and promoter methylation as a candidate biomarker in gastric cancer. Oncol. Lett. 2018, 15, 8833–8840.
- Jonkers, J.; Pai, P.; Sukumar, S. Multiple roles of HOX proteins in Metastasis: Let me count the ways. Cancer Metastasis Rev. 2020, 39, 661–679.
- Li, B.; Huang, Q.; Wei, G.H. The Role of HOX Transcription Factors in Cancer Predisposition and Progression. Cancers 2019, 11, 528.
- Sun, Y.; Zhou, B.; Mao, F.; Xu, J.; Miao, H.; Zou, Z.; Phuc Khoa, L.T.; Jang, Y.; Cai, S.; Witkin, M.; et al. HOXA9 Reprograms the Enhancer Landscape to Promote Leukemogenesis. Cancer Cell 2018, 34, 643–658.e5.
- De Benedetti, A.; Graff, J.R. eIF-4E expression and its role in malignancies and metastases. Oncogene 2004, 23, 3189–3199.
- Cillo, C.; Schiavo, G.; Cantile, M.; Bihl, M.P.; Sorrentino, P.; Carafa, V.; D’Armiento, M.; Roncalli, M.; Sansano, S.; Vecchione, R.; et al. The HOX gene network in hepatocellular carcinoma. Int. J. Cancer 2011, 129, 2577–2587.
- Guerra, S.L.; Maertens, O.; Kuzmickas, R.; De Raedt, T.; Adeyemi, R.O.; Guild, C.J.; Guillemette, S.; Redig, A.J.; Chambers, E.S.; Xu, M.; et al. A Deregulated HOX Gene Axis Confers an Epigenetic Vulnerability in KRAS-Mutant Lung Cancers. Cancer Cell 2020, 37, 705–719.e706.
- Yuan, Y.; Sun, S.; Jiao, N.; Shu, Y.; Zhang, Y. Upregulation of HOXA10 Protein Expression Predicts Poor Prognosis for Colorectal Cancer. Genet. Test. Mol. Biomark. 2018, 22, 390–397.
- Adwan, H.; Zhivkova-Galunska, M.; Georges, R.; Eyol, E.; Kleeff, J.; Giese, N.A.; Friess, H.; Bergmann, F.; Berger, M.R. Expression of HOXC8 is inversely related to the progression and metastasis of pancreatic ductal adenocarcinoma. Br. J. Cancer 2011, 105, 288–295.
- Lu‐Yan Shen; Ting Zhou; Ya‐Bing Du; Qi Shi; Ke-Neng Chen; Targeting HOX/PBX dimer formation as a potential therapeutic option in esophageal squamous cell carcinoma. Cancer Science 2019, 110, 1735-1745, 10.1111/cas.13993.
- Ting Zhou; Hao Fu; Bin Dong; Liang Dai; Yongbo Yang; Wanpu Yan; Luyan Shen; HOXB7 mediates cisplatin resistance in esophageal squamous cell carcinoma through involvement of DNA damage repair. Thoracic Cancer 2019, 11, 3071-3085, 10.1111/1759-7714.13142.
- Richard Morgan; Patricia Macanas Pirard; Liesl Shears; Jastinder Sohal; Ruth Pettengell; Hardev S. Pandha; Antagonism of HOX/PBX Dimer Formation Blocks the In vivo Proliferation of Melanoma. Cancer Research 2007, 67, 5806-5813, 10.1158/0008-5472.can-06-4231.
- M. Cristina Errico; Federica Felicetti; Lisabianca Bottero; Gianfranco Mattia; Alessandra Boe; Nadia Felli; Marina Petrini; Maria Bellenghi; Hardev S. Pandha; Marco Calvaruso; et al. The abrogation of the HOXB7/PBX2 complex induces apoptosis in melanoma through the miR-221&222-c-FOS pathway. International Journal of Cancer 2013, 133, 879-892, 10.1002/ijc.28097.
- Richard Morgan; Lynn Plowright; Kevin J. Harrington; Agnieszka Michael; Hardev S. Pandha; Targeting HOX and PBX transcription factors in ovarian cancer. BMC Cancer 2010, 10, 89-89, 10.1186/1471-2407-10-89.
- Richard Morgan; Angie Boxall; Kevin J. Harrington; Guy R. Simpson; Cheryl Gillett; Agnieszka Michael; Hardev S. Pandha; Targeting the HOX/PBX dimer in breast cancer. Breast Cancer Research and Treatment 2012, 136, 389-398, 10.1007/s10549-012-2259-2.
- Hitoshi Ando; Atsushi Natsume; Takeshi Senga; Reiko Watanabe; Ichiro Ito; Masasuke Ohno; Kenichiro Iwami; Fumiharu Ohka; Kazuya Motomura; Sayano Kinjo; et al. Peptide-based inhibition of the HOXA9/PBX interaction retards the growth of human meningioma. Cancer Chemotherapy and Pharmacology 2013, 73, 53-60, 10.1007/s00280-013-2316-5.
- Richard Morgan; Angie Boxall; Kevin J. Harrington; Guy R Simpson; Agnieszka Michael; Hardev S. Pandha; Targeting HOX transcription factors in prostate cancer. BMC Urology 2014, 14, 17-17, 10.1186/1471-2490-14-17.
- Zejuan Li; Zhiyu Zhang; Yuanyuan Li; Stephen Arnovitz; Ping Chen; Hao Huang; X. Jiang; Gia-Ming Hong; Rejani B. Kunjamma; Haomin Ren; et al. PBX3 is an important cofactor of HOXA9 in leukemogenesis. Blood 2013, 121, 1422-1431, 10.1182/blood-2012-07-442004.
- Vivek S. Chopra; Rakesh K. Mishra; “Mir”acles in hox gene regulation. BioEssays 2006, 28, 445-448, 10.1002/bies.20401.
- Rajnish A. Gupta; Nilay Shah; Kevin C. Wang; Jeewon Kim; Hugo M. Horlings; David J. Wong; Miao-Chih Tsai; Tiffany Hung; Pedram Argani; John L. Rinn; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071-1076, 10.1038/nature08975.
- Karl Agger; Paul A. C. Cloos; Jesper Christensen; Diego Pasini; Simon Rose; Juri Rappsilber; Irina Issaeva; Eli Canaani; Anna Elisabetta Salcini; Kristian Helin; et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 2007, 449, 731-734, 10.1038/nature06145.
- Morgan, R.; El-Tanani, M.; Hunter, K.D.; Harrington, K.J.; Pandha, H.S. Targeting HOX/PBX dimers in cancer. Oncotarget 2017, 8, 32322–32331.