Scaffold proteins are typically thought of as multi-domain “bridging molecules.” They serve
as crucial regulators of key signaling events by simultaneously binding multiple participants involved
in specific signaling pathways. In the case of epidermal growth factor (EGF)-epidermal growth factor
receptor (EGFR) binding, the activated EGFR contacts cytosolic SRC tyrosine-kinase, which then
becomes activated. This process leads to the phosphorylation of SRC-substrates, including the tyrosine
kinase substrates (TKS) scaffold proteins. The TKS proteins serve as a platform for the recruitment of
key players in EGFR signal transduction, promoting cell spreading and migration. The TKS4 and the
TKS5 scaffold proteins are tyrosine kinase substrates with four or five SH3 domains, respectively.
Their structural features allow them to recruit and bind a variety of signaling proteins and to anchor
them to the cytoplasmic surface of the cell membrane.
TKS4 and TKS5 had been recognized for their involvement in cellular motility, reactive oxygen species-dependent processes,
and embryonic development. Furthermore, TKS4 has also been implicated in the regulation of homeostasis of
mature adipose and bone tissue.
- scaffold protein
- tyrosine kinase substrates
- TKS4
- TKS5
- Beige adipose tissue
- osteoporosis
- EMT
- EGFR
- Src
- SH3 domai
- adaptor protein
- mesenchymal stem cells
- MSC
- FTHS
- podosome
- invadopodia
- rare hereditary disease
- Frank-ter Haar syndrome
- OMIM:249420
- Tks4-KO mice
- SH
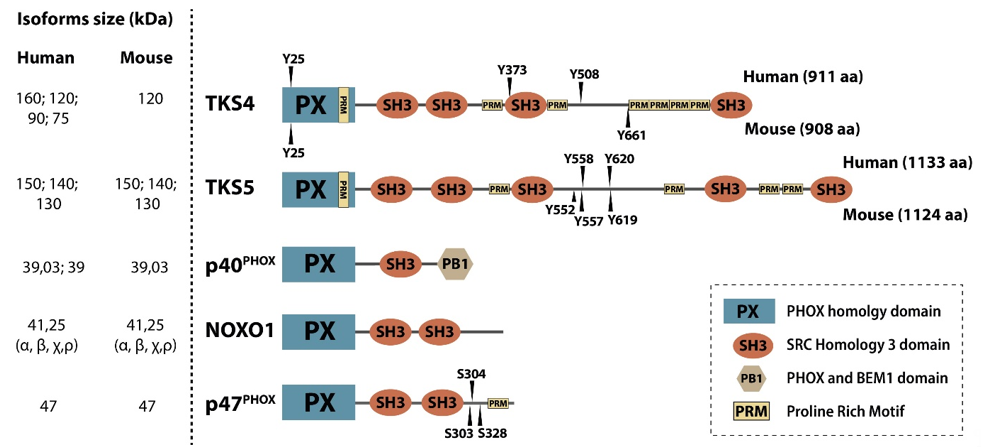
Scaffold proteins modulate intracellular signaling by bringing regulatory proteins, enzymes, or cytoskeletal structures in close proximity [1]. TKS molecules are large scaffold proteins earning their name from the early observation that they serve as tyrosine kinase substrates of SRC kinase [2–4]. TKS4 and TKS5 contain one Phox Homology (PX) domain, conserved linear motifs, e.g., several proline-rich motifs (PRMs), and four or five SRC Homology 3 (SH3) domains, respectively. Other names for TKS5 are SH3 and PX domain-containing protein 2A (SH3PXD2A) and Five SH3 domains (FISH), while TKS4 is also known as SH3 and PX domain-containing protein 2B (SH3PXD2B), Homolog of FISH (HOFI), and a factor of adipocyte differentiation 49 (Fad49), reflecting some of their known characteristics [3,5]. The main function of the PX domain is to link the TKS scaffold proteins to the cell membrane via phosphoinositide binding [2,6]. The SH3 domains serve as docking sites for signaling molecules and mediate protein-protein interactions [7]. It is likely that the PRMs of the TKS proteins represent contact sites for SH3 domain-containing molecules (Figure 1). The TKS proteins are phylogenetically related and are expressed in vertebrates, and TKS-like genes are widely present in invertebrates [8]. TKS scaffold proteins are broadly expressed in tissues except for the testis for TKS4, and the spleen and testis for TKS5 [2,3]. They are also expressed in several transformed cell lines [2,9].
Figure 1. Members of the p47 organizer protein family. The p47 organizer family consists of five structurally similar adaptor/scaffold proteins containing an N-terminal PX domain followed by several SH3 domains, namely p40phox, NOXO1, p47phox, TKS4, and TKS5. Experimentally confirmed SRC kinase tyrosine phosphorylation sites (“Y”) in the human and mouse TKS proteins are shown above and below the depicted domain architecture, respectively.
TKS4 and TKS5 Affect Multiple Biological Processes from Growth Factor Receptor Signaling to Metastasis to Homeostasis of Adipose and Bone Tissue
EGFR Signaling via TKS4 and TKS5
Receptor tyrosine kinases (RTK) are transmembrane proteins that control several cellular processes, ranging from proliferation to differentiation and cell migration. Following the binding of their extracellular ligands, RTKs dimerize, undergo auto-phosphorylation on multiple tyrosine residues in their cytoplasmic region, and associate with intracellular signaling molecules. Diverse molecular cascades transmit the signal from RTKs to their final effector molecules, ultimately leading to the modulation of distinct biological processes within the cell [20].
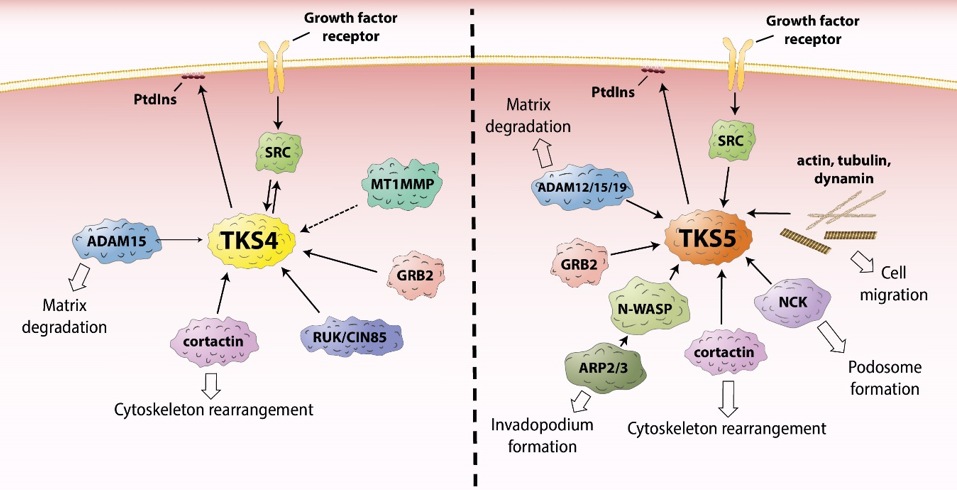
Epidermal growth factor receptor (EGFR) is one of the most well-studied RTKS. Upon activation, it initiates several signal transduction cascades, including the RAS-RAF-MEK, phosphatidylinositol 3 (PI3)- kinase-AKT, PLCγ, and JAK-STAT pathways [21]. Moreover, active EGFR binds cytosolic SRC tyrosine-kinase, which then becomes activated [22–25]. This process leads to the phosphorylation of SRC-substrates, including the TKS scaffold proteins, which are known to be involved in EGFR signaling [11,26,27]. The TKS proteins serve as a platform for the recruitment of key players in EGFR signal transduction (Figure 2, Table 1), promoting cell spreading and migration [9,11,28–30]. In response to EGFR activation, PI3 kinases are activated, and lipids are phosphorylated in the plasma membrane. For example, phosphatidylinositol (4,5)-bisphosphate (PI(4,5)P2) is converted to phosphatidylinositol (3,4,5)-trisphosphate (PI(3,4,5)P3) [31]. According to a model proposed by Bögel et al., the phosphorylated lipid residues anchor the PX domain of TKS4 and translocate the scaffold protein from the cytoplasm to the plasma membrane [11]. On the other arm of the signaling pathway, SRC kinase is also activated by binding to the intracellular tail of EGFR [22–25] subsequently phosphorylating tyrosine residues on TKS4 (i.e., Tyr25, Tyr373, Tyr508) (Figure 1) [2]. Phosphorylated TKS4 can bind activated SRC by interacting with both its SH2 and SH3 domains. In this complex, SRC remains active for a prolonged period of time and may phosphorylate multiple downstream molecules/partners [32]. This direct interaction between TKS4 with SRC was shown to involve the proline-rich region PSRPLPDAP (residues 466–474) and the tyrosine-phosphorylated pYEEI motif (residues 508–511) of TKS4 (both located between the third and fourth SH3 domains) and the SH3 and SH2 domains of SRC, respectively [32]. Upon PI3 kinase activation, TKS5 also translocates to the plasma membrane in epidermal growth factor (EGF)-stimulated cells [26]. The PX domain of both TKS4 and 5 was found to be essential for the participation of the molecules in EGFR signaling and for the phosphorylation of TKS4 and 5 by activated SRC [11,26]. TKS4 forms a complex with EGFR in which either SRC or a yet unidentified protein may serve as a bridge between the two molecules [11,32]. For example, growth factor receptor binding protein 2 (GRB2) has been identified as a binding partner of both EGFR and TKS4 [28]. No strong interaction between TKS5 and EGFR or SRC has been detected so far, suggesting that, despite their structural similarities, there is only a partial overlap between the regulation of TKS4 and TKS5 in EGF signaling [11,26].
Figure 2. The role of TKS proteins in the recruitment of signaling molecules. ADAM12/15/19 – a disintegrin and metalloprotease 12/15/19, ARP2/3 – actin-related protein 2/3, GRB2 – growth factor receptor binding protein 2, MT1MMP – membrane type 1 matrix metalloprotease, NCK – non-catalytic region of tyrosine kinase adaptor protein, N-WASP – neural Wiskott-Aldrich syndrome protein, PtdIns – phosphatidylinositol, RUK/CIN85 - regulator of ubiquitous kinase/Cbl-interacting protein of 85 kDa, SRC - proto-oncogene tyrosine-protein kinase Src.
Table 1. Known protein binding partners of TKS4 and TKS5. The known binding partners of (a) TKS4 and (b) TKS5 are shown with the methods of detection and the binding sites within the TKS molecules. Some of the well-described functions of the binding partners are also listed. ECM – extracellular matrix, EMT – epithelial-mesenchymal transition, ITC – isothermal titration calorimetry, NOX1 – NADPH oxidase 1, PRR – proline-rich region, ROS – reactive oxygen species, RTK – receptor tyrosine kinase. * The first and second SH3 domains cooperate to form a common “super SH3 platform” and allow the binding of the proline-rich region of the partner protein [33].
Table 1a.
|
TKS4 |
|||
|
Partner |
Method |
TKS4-Interacting Site |
Function |
|
ADAM15 [34] |
GST pull-down assay |
4th SH3 domain |
Ectodomain shedding, cell adhesion, and signaling [35] |
|
Cortactin [9] |
Co-immunoprecipitation, GST pull-down assay, immunofluorescence co-localization |
Unknown |
Regulation of actin cytoskeleton [36] |
|
CR16 [37] |
GST pull-down assay |
Weak interaction with the 2nd, 3rd, and 4th SH3 domains |
Reorganization of actin cytoskeleton [38] |
|
DNM2 [37] |
GST pull-down assay |
3rd SH3 domain |
Endo-/exocytosis [37] |
|
FasL (CD178) [39] |
Phage display screening |
3rd and 4th SH3 domains |
Apoptosis induction [40] |
|
GRB2 [28] |
Affinity purification–selected reaction monitoring mass spectrometry |
Unknown |
Adaptor protein involved in the regulation of RTK signaling, cycle progression, actin-based cell motility, podosome formation [41] |
|
NOXA1 [42,43] |
Co-immunoprecipitation, GST pull-down assay |
Unknown |
ROS generation through NOX1 activation [44] |
|
N-WASP [37] |
GST pull-down assay |
2nd SH3 domain |
A scaffold protein regulating actin cytoskeleton reorganization, and actin polymerization during cell motility and invasion [45] |
|
OPHN1 [37] |
GST pull-down assay |
3rd SH3 domain |
Endo-/exocytosis [37] |
|
RUK/CIN85 [46] |
GST pull-down assay |
Unknown |
Adaptor protein that recruits endocytotic regulatory proteins, and regulates RTK internalization, trafficking, and degradation [47] |
|
SRC [9,32] |
Co-immunoprecipitation; GST pull-down and fluorescence-polaziation assays, Duolink proximity ligation assay |
PRR (aa: 466–474); P-Tyr motif (aa: 508–511) |
Regulation of cell growth, differentiation, proliferation, survival, adhesion, migration, and motility [9,32] |
|
SYNJ1 [37] |
GST pull-down assay |
3rd SH3 domain and weak interaction with the 4th SH3 domain |
Endo-/exocytosis [37] |
Table 1b.
|
TKS5 |
|||
|
Partner |
Method |
TKS5-Interacting Site |
Function |
|
ADAM12 [6] |
Co-immunoprecipitation, immunofluorescence co-localization |
5th SH3 domain |
Cell adhesion and fusion, extracellular matrix restructuring, reorganization of actin cytoskeleton, regulation of ectodomain shedding [48] |
|
ADAM15 [6] |
Co-immunoprecipitation |
5th SH3 domain |
Cell adhesion, degradation of ECM components, ectodomain shedding of membrane-bound growth factors [35] |
|
ADAM19 [6] |
Phage display screen, co-immunoprecipitation |
5th SH3 domain |
Extracellular matrix breakdown and reconstruction, ectodomain shedding, role in embryogenesis, cardiovascular system development, obesity, and insulin resistance [49] |
|
b-dystroglycan [50] |
Phage display screen, GST pull-down assay, co-immunoprecipitation, immunofluorescence co-localization |
3rd SH3 domain |
Links the extracellular matrix to the intracellular actin cytoskeleton [50] |
|
CircSKA3 [51] |
Co-immunoprecipitation, pull-down assay |
Not specified |
Circular RNA, an inducer of invadopodium formation [51] |
|
Drebrin [52] |
Co-immunoprecipitation |
Unknown |
An actin-binding protein involved in the regulation of actin filament organization, role in cell migration, cell process formation, intercellular communication, metastasis, and brain development [53] |
|
Dynamin [29,33] |
Peptide spot membrane assay, GST pull-down assay, ITC, immunofluorescence co-localization, GST pull-down assay, mass spectrometry/Western blotting |
1st and 2nd SH3 domains; 1st and 5th SH3 domains |
Regulation of actin cytoskeleton, podosome/invadopodium formation, role in endocytosis [54] |
|
F-actin [29] |
GST pull-down assay, and mass spectrometry |
5th SH3 domain |
Component of cytoskeleton [55] |
|
FasL (CD178) [39] |
Phage display screening |
5th SH3 domain |
Apoptosis induction [40] |
|
FGD1 [56] |
Co-immunoprecipitation and mass spectrometry, GST pull-down assay, immunofluorescence co-localization |
4th and 5th SH3 domains |
A guanine nucleotide exchange factor for the Rho-GTPase CDC42, assembly of podosomes and invadopodia, control of secretory membrane-trafficking, and cell cycle [56,57] |
|
Girdin [58] |
Co-immunoprecipitation, immunofluorescence co-localization |
Unknown |
actin-binding protein regulating actin remodeling and cell polarity, collective migration of neuroblasts, epithelial and cancer cells [59] |
|
GRB2 [28,29] |
Co-immunoprecipitation |
Polyproline sequences |
An adaptor protein involved in cell cycle progression and actin-based cell motility, podosome formation [41] |
|
IRTKS [60] |
GST pull-down assay |
First binding site located in the segment comprising the 1st and 2nd SH3 domains, second binding site located in the segment comprising the 3rd and 4th SH3 domains |
Regulation of plasma membrane dynamics, actin cytoskeleton remodeling, cell migration and polarization, insulin signaling [61] |
|
MT4-MMP [62] |
Co-immunoprecipitation |
Unknown |
Induction of invadopodia and amoeboid movement, degradation of ECM components, role in hypoxia-mediated metastasis [62] |
|
NCK [52] |
Co-immunoprecipitation, fluorescence co-localization |
Linker region between the 3rd and 4th SH3 domains containing pY557 |
Adaptor protein involved in cytoskeletal remodeling, invadopodium formation, cell proliferation [63] |
|
Nogo-B [29] |
GST pull-down assay and mass spectrometry |
5th SH3 domain |
Roles in vascular remodeling, cell migration and proliferation, and EMT [64] |
|
NOXA1 [42,43] |
Co-immunoprecipitation, GST pull-down assay |
One or more of the five SH3 domains |
ROS generation through NOX1 activation [44] |
|
N-WASP [29] |
GST pull-down assay and mass spectrometry/Western blotting, co-immunoprecipitation |
All five SH3 domains |
A scaffold protein regulating actin cytoskeleton reorganization, and actin polymerization during cell motility and invasion [45] |
|
p22phox [65] |
Co-immunoprecipitation |
1st and 2nd SH3 domains |
Subunit of NADPH oxidases involved in ROS generation through NOX activity [66] |
|
Rab40b [67] |
GST pull-down assay, co-immunoprecipitation, |
PX-domain: sites 14-KRR-19 and Y24 in 23-YVYI-28 |
A GTPase required for the sorting and secretion of MMP2 and MMP9, promotion of migration, invasion, and metastasis of cancer cells [67,68] |
|
RET [69] |
Co-immunoprecipitation, immunofluorescence co-localization |
Unknown |
A receptor tyrosine kinase mediating stress fiber formation, cell polarization, directional migration and invasion, enhancement of proteolytic activity [69] |
|
SOS1 [33] |
Immunofluorescence co-localization, peptide spot membrane assay, GST pull-down assay, isothermal titration calorimetry |
1st and 2nd SH3 domains * |
A guanine nucleotide exchange factor promoting Ras and Rac activation downstream of a variety of receptors such as RTKs [70] |
|
Tubulin [29] |
GST pull-down assay and mass spectrometry |
3rd SH3 domain |
Component of microtubules, affects cell division, differentiation, |
|
WIP [29] |
GST pull-down assay and mass spectrometry |
3rd and 5th SH3 domains |
Regulation of actin cytoskeleton assembly and remodeling [72] |
|
XB130 [73] |
Yeast two-hybrid screening, co-immunoprecipitation, GST pull-down assay, immunofluorescence co-localization |
5th SH3 domain |
A scaffold protein influencing cell growth, survival, and migration [73] |
|
Zyxin [29] |
GST pull-down assay and mass spectrometry |
3rd and 5th SH3 domains |
A focal adhesion protein involved in actin cytoskeleton assembly [74] |
In recent years, more possible interaction partners of TKS4 have been identified (Table 1(A)). One possible partner is cortactin [9], a well-known substrate of SRC localized to cortical actin structures within cells. Cortactin can bind the actin-related protein-2/3 (ARP2/3) and neural Wiskott-Aldrich syndrome protein (N-WASP) proteins, and it mediates actin polymerization [75–77]. Therefore, TKS4 was expected to be involved in EGFR signaling-mediated actin cytoskeleton assembly and rearrangement. This proposed mechanism was confirmed by Lányi et al. [9]. They found that, in response to EGF stimulation, TKS4 associates with cellular motility-associated membrane ruffles. They also showed that, when constitutively active SRC is present, TKS4 accumulates in podosomes (actin-rich membrane protrusions involved in cell motility, see below) while forming a complex with SRC and cortactin [9]. TKS5 has also been reported to bind cortactin and other proteins important in the regulation of actin cytoskeleton assembly, including N-WASP, non-catalytic region of tyrosine kinase adaptor protein (NCK), and GRB2 (for a full list, see Table 1b) [10,29,52,78,79].
Long version of the text: https://doi.org/10.3390/ijms21218117
Advances in Understanding TKS4 and TKS5: Molecular Scaffolds Regulating Cellular Processes from Podosome and Invadopodium Formation to Differentiation and Tissue Homeostasis
Int. J. Mol. Sci. 2020, 21(21), 8117
References
- Buday, L.; Tompa, P. Functional classification of scaffold proteins and related molecules. FEBS J. 2010, 277, 4348–4355, doi:10.1111/j.1742-4658.2010.07864.x.
- Buschman, M.D.; Bromann, P.A.; Cejudo-Martin, P.; Wen, F.; Pass, I.; Courtneidge, S.A. The Novel Adaptor Protein Tks4 (SH3PXD2B) is Required for Functional Podosome Formation. Biol. Cell 2009, 20, 1302–1311, doi:10.1091/mbc.e08-09-0949.
- Lock, P.; Abram, C.L.; Gibson, T.; Courtneidge, S.A. A new method for isolating tyrosine kinase substrates used to identify Fish, an SH3 and PX domain-containing protein, and Src substrate. EMBO J. 1998, 17, 4346–4357, doi:10.1093/emboj/17.15.4346.
- Weaver, A.M. Regulation of Cancer Invasion by Reactive Oxygen Species and Tks Family Scaffold Proteins. Signal. 2009, 2, pe56–pe56, doi:10.1126/scisignal.288pe56.
- Ádám, C.; Fekete, A.; Bőgel, G.; Németh, Z.; Tőkési, N.; Ovádi, J.; Liliom, K.; Pesti, S.; Geiszt, M.; Buday, L. Accumulation of the PX domain mutant Frank-ter Haar syndrome protein Tks4 in aggresomes. Cell Commun. Signal. 2015, 13, 33, doi:10.1186/s12964-015-0108-8.
- Abram, C.L.; Seals, D.F.; Pass, I.; Salinsky, D.; Maurer, L.; Roth, T.M.; Courtneidge, S.A. The Adaptor Protein Fish Associates with Members of the ADAMs Family and Localizes to Podosomes of Src-transformed Cells. Biol. Chem. 2003, 278, 16844–16851, doi:10.1074/jbc.M300267200.
- Kurochkina, N.; Guha, U. SH3 domains: Modules of protein–protein interactions. Rev. 2013, 5, 29–39, doi:10.1007/s12551-012-0081-z.
- Kawahara, T.; Lambeth, J.D. Molecular evolution of Phox-related regulatory subunits for NADPH oxidase enzymes. BMC Evol. Biol. 2007, 7, 178, doi:10.1186/1471-2148-7-178.
- Lányi, Á.; Baráth, M.; Péterfi, Z.; Bőgel, G.; Orient, A.; Simon, T.; Petrovszki, E.; Kis-Tóth, K.; Sirokmány, G.; Rajnavölgyi, É.; et al. The Homolog of the Five SH3-Domain Protein (HOFI/SH3PXD2B) Regulates Lamellipodia Formation and Cell Spreading. PLoS ONE 2011, 6, e23653, doi:10.1371/journal.pone.0023653.
- Daly, C.; Logan, B.; Breeyear, J.; Whitaker, K.; Ahmed, M.; Seals, D.F. Tks5 SH3 domains exhibit differential effects on invadopodia development. PLoS ONE 2020, 15, e0227855, doi:10.1371/journal.pone.0227855.
- Bögel, G.; Gujdár, A.; Geiszt, M.; Lányi, Á.; Fekete, A.; Sipeki, S.; Downward, J.; Buday, L. Frank-ter Haar Syndrome Protein Tks4 Regulates Epidermal Growth Factor-dependent Cell Migration. Biol. Chem. 2012, 287, 31321–31329, doi:10.1074/jbc.M111.324897.
- Hishida, T.; Eguchi, T.; Osada, S.; Nishizuka, M.; Imagawa, M. A novel gene, fad49, plays a crucial role in the immediate early stage of adipocyte differentiation via involvement in mitotic clonal expansion. FEBS J. 2008, 275, 5576–5588, doi:10.1111/j.1742-4658.2008.06682.x.
- Teasdale, R.D.; Collins, B.M. Insights into the PX (phox-homology) domain and SNX (sorting nexin) protein families: Structures, functions and roles in disease. J. 2012, 441, 39–59, doi:10.1042/BJ20111226.
- Yaffe, M.B. The p47phox PX Domain: Two Heads Are Better Than One! Structure 2002, 10, 1288–1290, doi:10.1016/S0969-2126(02)00860-2.
- Groemping, Y.; Lapouge, K.; Smerdon, S.J.; Rittinger, K. Molecular Basis of Phosphorylation-Induced Activation of the NADPH Oxidase. Cell 2003, 113, 343–355, doi:10.1016/S0092-8674(03)00314-3.
- Belambri, S.A.; Rolas, L.; Raad, H.; Hurtado-Nedelec, M.; Dang, P.M.-C.; El-Benna, J. NADPH oxidase activation in neutrophils: Role of the phosphorylation of its subunits. J. Clin. Investig. 2018, 48, e12951, doi:10.1111/eci.12951.
- El-Benna, J.; Dang, P.M.-C.; Gougerot-Pocidalo, M.-A.; Marie, J.-C.; Braut-Boucher, F. p47phox, the phagocyte NADPH oxidase/NOX2 organizer: Structure, phosphorylation and implication in diseases. Mol. Med. 2009, 41, 217–225, doi:10.3858/emm.2009.41.4.058.
- Meijles, D.N.; Fan, L.M.; Howlin, B.J.; Li, J.-M. Molecular Insights of p47phox Phosphorylation Dynamics in the Regulation of NADPH Oxidase Activation and Superoxide Production. Biol. Chem. 2014, 289, 22759–22770, doi:10.1074/jbc.M114.561159.
- Oikawa, T.; Oyama, M.; Kozuka-Hata, H.; Uehara, S.; Udagawa, N.; Saya, H.; Matsuo, K. Tks5-dependent formation of circumferential podosomes/invadopodia mediates cell–cell fusion. Cell Biol. 2012, 197, 553–568, doi:10.1083/jcb.201111116.
- Casaletto, J.B.; McClatchey, A.I. Spatial regulation of receptor tyrosine kinases in development and cancer. Rev. Cancer 2012, 12, 387–400, doi:10.1038/nrc3277.
- Wee, P.; Wang, Z. Epidermal Growth Factor Receptor Cell Proliferation Signaling Pathways. Cancers 2017, 9, 52, doi:10.3390/cancers9050052.
- Belsches, A.P.; Haskell, M.D.; Parsons, S.J. Role of c-Src tyrosine kinase in EGF-induced mitogenesis. Biosci. 1997, 2, d501–d518, doi:10.2741/a208.
- Alonso, G.; Koegl, M.; Mazurenko, N.; Courtneidge, S.A. Sequence Requirements for Binding of Src Family Tyrosine Kinases to Activated Growth Factor Receptors. Biol. Chem. 1995, 270, 9840–9848, doi:10.1074/jbc.270.17.9840.
- Bromann, P.A.; Korkaya, H.; Courtneidge, S.A. The interplay between Src family kinases and receptor tyrosine kinases. Oncogene 2004, 23, 7957–7968, doi:10.1038/sj.onc.1208079.
- Sierke, S.L.; Longo, G.M.; Koland, J.G. Structural Basis of Interactions Between Epidermal Growth Factor Receptor and SH2 Domain Proteins. Biophys. Res. Commun. 1993, 191, 45–54, doi:10.1006/bbrc.1993.1182.
- Fekete, A.; Bőgel, G.; Pesti, S.; Péterfi, Z.; Geiszt, M.; Buday, L. EGF regulates tyrosine phosphorylation and membrane-translocation of the scaffold protein Tks5. Mol. Signal. 2013, 8, 8, doi:10.1186/1750-2187-8-8.
- Gianni, D.; Taulet, N.; DerMardirossian, C.; Bokoch, G.M. c-Src–Mediated Phosphorylation of NoxA1 and Tks4 Induces the Reactive Oxygen Species (ROS)–Dependent Formation of Functional Invadopodia in Human Colon Cancer Cells. Biol. Cell 2010, 21, 4287–4298, doi:10.1091/mbc.e10-08-0685.
- Bisson, N.; James, D.A.; Ivosev, G.; Tate, S.A.; Bonner, R.; Taylor, L.; Pawson, T. Selected reaction monitoring mass spectrometry reveals the dynamics of signaling through the GRB2 adaptor. Biotechnol. 2011, 29, 653–658, doi:10.1038/nbt.1905.
- Oikawa, T.; Itoh, T.; Takenawa, T. Sequential signals toward podosome formation in NIH-src cells. Cell Biol. 2008, 182, 157–169, doi:10.1083/jcb.200801042.
- Crimaldi, L.; Courtneidge, S.A.; Gimona, M. Tks5 recruits AFAP-110, p190RhoGAP, and cortactin for podosome formation. Cell Res. 2009, 315, 2581–2592, doi:10.1016/j.yexcr.2009.06.012.
- Thapa, N.; Tan, X.; Choi, S.; Lambert, P.F.; Rapraeger, A.C.; Anderson, R.A. The Hidden Conundrum of Phosphoinositide Signaling in Cancer. Trends Cancer 2016, 2, 378–390, doi:10.1016/j.trecan.2016.05.009.
- Dülk, M.; Szeder, B.; Glatz, G.; Merő, B.L.; Koprivanacz, K.; Kudlik, G.; Vas, V.; Sipeki, S.; Cserkaszky, A.; Radnai, L.; et al. EGF Regulates the Interaction of Tks4 with Src through Its SH2 and SH3 Domains. Biochemistry 2018, 57, 4186–4196, doi:10.1021/acs.biochem.8b00084.
- Rufer, A.C.; Rumpf, J.; von Holleben, M.; Beer, S.; Rittinger, K.; Groemping, Y. Isoform-Selective Interaction of the Adaptor Protein Tks5/FISH with Sos1 and Dynamins. Mol. Biol. 2009, 390, 939–950, doi:10.1016/j.jmb.2009.05.025.
- Mao, M.; Thedens, D.R.; Chang, B.; Harris, B.S.; Zheng, Q.Y.; Johnson, K.R.; Donahue, L.R.; Anderson, M.G. The podosomal-adaptor protein SH3PXD2B is essential for normal postnatal development. Genome 2009, 20, 462, doi:10.1007/s00335-009-9210-9.
- Mattern, J.; Roghi, C.S.; Hurtz, M.; Knäuper, V.; Edwards, D.R.; Poghosyan, Z. ADAM15 mediates upregulation of Claudin-1 expression in breast cancer cells. Rep. 2019, 9, 12540, doi:10.1038/s41598-019-49021-3.
- Sharafutdinov, I.; Backert, S.; Tegtmeyer, N. Cortactin: A Major Cellular Target of the Gastric Carcinogen Helicobacter pylori. Cancers 2020, 12, 159, doi:10.3390/cancers12010159.
- Kropyvko, S.V. New partners of TKS4 scaffold protein. Cell 2015, 31, 395–401, doi:10.7124/bc.0008FC.
- Kropyvko, S.; Gryaznova, T.; Morderer, D.; Rynditch, A. Mammalian verprolin CR16 acts as a modulator of ITSN scaffold proteins association with actin. Biophys. Res. Commun. 2017, 484, 813–819, doi:10.1016/j.bbrc.2017.01.177.
- Voss, M.; Lettau, M.; Janssen, O. Identification of SH3 domain interaction partners of human FasL (CD178) by phage display screening. BMC Immunol. 2009, 10, 53, doi:10.1186/1471-2172-10-53.
- Glukhova, X.A.; Trizna, J.A.; Proussakova, O.V.; Gogvadze, V.; Beletsky, I.P. Impairment of Fas-ligand–caveolin-1 interaction inhibits Fas-ligand translocation to rafts and Fas-ligand-induced cell death. Cell Death Dis. 2018, 9, 1–12, doi:10.1038/s41419-017-0109-1.
- Giubellino, A.; Burke, J.T.R.; Bottaro, D.P. Grb2 signaling in cell motility and cancer. Expert Opin. Ther. Targets 2008, 12, 1021–1033, doi:10.1517/14728222.12.8.1021.
- Gianni, D.; Diaz, B.; Taulet, N.; Fowler, B.; Courtneidge, S.A.; Bokoch, G.M. Novel p47phox-Related Organizers Regulate Localized NADPH Oxidase 1 (Nox1) Activity. Signal. 2009, 2, ra54, doi:10.1126/scisignal.2000370.
- Gianni, D.; DerMardirossian, C.; Bokoch, G.M. Direct interaction between Tks proteins and the N-terminal proline-rich region (PRR) of NoxA1 mediates Nox1-dependent ROS generation. J. Cell Biol. 2011, 90, 164–171, doi:10.1016/j.ejcb.2010.05.007.
- Schröder, K.; Weissmann, N.; Brandes, R.P. Organizers and activators: Cytosolic Nox proteins impacting on vascular function. Free Radic. Biol. Med. 2017, 109, 22–32, doi:10.1016/j.freeradbiomed.2017.03.017.
- Hou, J.; Yang, H.; Huang, X.; Leng, X.; Zhou, F.; Xie, C.; Zhou, Y.; Xu, Y. N-WASP promotes invasion and migration of cervical cancer cells through regulating p38 MAPKs signaling pathway. J. Transl. Res. 2017, 9, 403–415.
- Bazalii, A.V.; Samoylenko, A.A.; Petukhov, D.M.; Rynditch, A.V.; Redowicz, M.-J.; Drobot, L.B. Interaction between adaptor proteins Ruk/CIN85 and Tks4 in normal and tumor cells of different tissue origins. Cell 2014, 30, 37–41, doi:10.7124/bc.00087A.
- Kong, M.S.; Hashimoto-Tane, A.; Kawashima, Y.; Sakuma, M.; Yokosuka, T.; Kometani, K.; Onishi, R.; Carpino, N.; Ohara, O.; Kurosaki, T.; et al. Inhibition of T cell activation and function by the adaptor protein CIN85. Signal. 2019, 12, doi:10.1126/scisignal.aav4373.
- Nyren-Erickson, E.K.; Jones, J.M.; Srivastava, D.K.; Mallik, S. A disintegrin and metalloproteinase-12 (ADAM12): Function, roles in disease progression, and clinical implications. Biophys. Acta BBA Gen. Subj. 2013, 1830, 4445–4455, doi:10.1016/j.bbagen.2013.05.011.
- Weerasekera, L.; Rudnicka, C.; Sang, Q.-X.; Curran, J.E.; Johnson, M.P.; Moses, E.K.; Göring, H.H.H.; Blangero, J.; Hricova, J.; Schlaich, M.; et al. ADAM19: A Novel Target for Metabolic Syndrome in Humans and Mice. Inflamm. 2017, 2017, doi:10.1155/2017/7281986.
- Thompson, O.; Kleino, I.; Crimaldi, L.; Gimona, M.; Saksela, K.; Winder, S.J. Dystroglycan, Tks5 and Src Mediated Assembly of Podosomes in Myoblasts. PLoS ONE 2008, 3, e3638, doi:10.1371/journal.pone.0003638.
- Du, W.W.; Yang, W.; Li, X.; Fang, L.; Wu, N.; Li, F.; Chen, Y.; He, Q.; Liu, E.; Yang, Z.; et al. The Circular RNA circSKA3 Binds Integrin β1 to Induce Invadopodium Formation Enhancing Breast Cancer Invasion. Ther. 2020, 28, 1287–1298, doi:10.1016/j.ymthe.2020.03.002.
- Stylli, S.S.; Stacey, T.T.I.; Verhagen, A.M.; Xu, S.S.; Pass, I.; Courtneidge, S.A.; Lock, P. Nck adaptor proteins link Tks5 to invadopodia actin regulation and ECM degradation. Cell Sci. 2009, 122, 2727–2740, doi:10.1242/jcs.046680.
- Shirao, T.; Hanamura, K.; Koganezawa, N.; Ishizuka, Y.; Yamazaki, H.; Sekino, Y. The role of drebrin in neurons. Neurochem. 2017, 141, 819–834, doi:10.1111/jnc.13988.
- Zhang, R.; Lee, D.M.; Jimah, J.R.; Gerassimov, N.; Yang, C.; Kim, S.; Luvsanjav, D.; Winkelman, J.; Mettlen, M.; Abrams, M.E.; et al. Dynamin regulates the dynamics and mechanical strength of the actin cytoskeleton as a multifilament actin-bundling protein. Cell Biol. 2020, 22, 674–688, doi:10.1038/s41556-020-0519-7.
- Grintsevich, E.E.; Yesilyurt, H.G.; Rich, S.K.; Hung, R.-J.; Terman, J.R.; Reisler, E. F-actin dismantling through a redox-driven synergy between Mical and cofilin. Cell Biol. 2016, 18, 876–885, doi:10.1038/ncb3390.
- Zagryazhskaya-Masson, A.; Monteiro, P.; Macé, A.-S.; Castagnino, A.; Ferrari, R.; Infante, E.; Duperray-Susini, A.; Dingli, F.; Lanyi, A.; Loew, D.; et al. Intersection of TKS5 and FGD1/CDC42 signaling cascades directs the formation of invadopodia. Cell Biol. 2020, 219, doi:10.1083/jcb.201910132.
- Genot, E.; Daubon, T.; Sorrentino, V.; Buccione, R. FGD1 as a central regulator of extracellular matrix remodelling—Lessons from faciogenital dysplasia. Cell Sci. 2012, 125, 3265–3270, doi:10.1242/jcs.093419.
- Ke, Y.; Bao, T.; Zhou, Q.; Wang, Y.; Ge, J.; Fu, B.; Wu, X.; Tang, H.; Shi, Z.; Lei, X.; et al. Discs large homolog 5 decreases formation and function of invadopodia in human hepatocellular carcinoma via Girdin and Tks5. J. Cancer 2017, 141, 364–376, doi:10.1002/ijc.30730.
- Wang, X.; Enomoto, A.; Weng, L.; Mizutani, Y.; Abudureyimu, S.; Esaki, N.; Tsuyuki, Y.; Chen, C.; Mii, S.; Asai, N.; et al. Girdin/GIV regulates collective cancer cell migration by controlling cell adhesion and cytoskeletal organization. Cancer Sci. 2018, 109, 3643–3656, doi:10.1111/cas.13795.
- Oikawa, T.; Matsuo, K. Possible role of IRTKS in Tks5-driven osteoclast fusion. Integr. Biol. 2012, 5, 511–515, doi:10.4161/cib.21252.
- Li, L.; Liu, H.; Baxter, S.S.; Gu, N.; Ji, M.; Zhan, X. The SH3 domain distinguishes the role of I-BAR proteins IRTKS and MIM in chemotactic response to serum. Biophys. Res. Commun. 2016, 479, 787–792, doi:10.1016/j.bbrc.2016.09.131.
- Yan, X.; Cao, N.; Chen, Y.; Lan, H.-Y.; Cha, J.-H.; Yang, W.-H.; Yang, M.-H. MT4-MMP promotes invadopodia formation and cell motility in FaDu head and neck cancer cells. Biophys. Res. Commun. 2020, 522, 1009–1014, doi:10.1016/j.bbrc.2019.12.009.
- Chaki, S.P.; Barhoumi, R.; Rivera, G.M. Nck adapter proteins promote podosome biogenesis facilitating extracellular matrix degradation and cancer invasion. Cancer Med. 2019, 8, 7385–7398, doi:10.1002/cam4.2640.
- Zhu, B.; Chen, S.; Hu, X.; Jin, X.; Le, Y.; Cao, L.; Yuan, Z.; Lin, Z.; Jiang, S.; Sun, L.; et al. Knockout of the Nogo-B Gene Attenuates Tumor Growth and Metastasis in Hepatocellular Carcinoma. Neoplasia 2017, 19, 583–593, doi:10.1016/j.neo.2017.02.007.
- Diaz, B.; Shani, G.; Pass, I.; Anderson, D.; Quintavalle, M.; Courtneidge, S.A. Tks5-Dependent, Nox-Mediated Generation of Reactive Oxygen Species Is Necessary for Invadopodia Formation. Signal. 2009, 2, ra53, doi:10.1126/scisignal.2000368.
- Nagaraj, C.; Tabeling, C.; Nagy, B.M.; Jain, P.P.; Marsh, L.M.; Papp, R.; Pienn, M.; Witzenrath, M.; Ghanim, B.; Klepetko, W.; et al. Hypoxic vascular response and ventilation/perfusion matching in end-stage COPD may depend on p22phox. Respir. J. 2017, 50, doi:10.1183/13993003.01651-2016.
- Jacob, A.; Linklater, E.; Bayless, B.A.; Lyons, T.; Prekeris, R. The role and regulation of Rab40b–Tks5 complex during invadopodia formation and cancer cell invasion. Cell Sci. 2016, 129, 4341–4353, doi:10.1242/jcs.193904.
- Li, Y.; Jia, Q.; Wang, Y.; Li, F.; Jia, Z.; Wan, Y. Rab40b upregulation correlates with the prognosis of gastric cancer by promoting migration, invasion, and metastasis. Oncol. 2015, 32, 126, doi:10.1007/s12032-015-0562-6.
- Lian, E.Y.; Hyndman, B.D.; Moodley, S.; Maritan, S.M.; Mulligan, L.M. RET isoforms contribute differentially to invasive processes in pancreatic ductal adenocarcinoma. Oncogene 2020, 1–18, doi:10.1038/s41388-020-01448-z.
- Gerboth, S.; Frittoli, E.; Palamidessi, A.; Baltanas, F.C.; Salek, M.; Rappsilber, J.; Giuliani, C.; Troglio, F.; Rolland, Y.; Pruneri, G.; et al. Phosphorylation of SOS1 on tyrosine 1196 promotes its RAC GEF activity and contributes to BCR-ABL leukemogenesis. Leukemia 2018, 32, 820–827, doi:10.1038/leu.2017.267.
- Prassanawar, S.S.; Panda, D. Tubulin heterogeneity regulates functions and dynamics of microtubules and plays a role in the development of drug resistance in cancer. J. 2019, 476, 1359–1376, doi:10.1042/BCJ20190123.
- Sokolik, C.G.; Qassem, N.; Chill, J.H. The Disordered Cellular Multi-Tasker WIP and Its Protein–Protein Interactions: A Structural View. Biomolecules 2020, 10, 1084, doi:10.3390/biom10071084.
- Moodley, S.; Hui Bai, X.; Kapus, A.; Yang, B.; Liu, M. XB130/Tks5 scaffold protein interaction regulates Src-mediated cell proliferation and survival. Biol. Cell 2015, 26, 4492–4502, doi:10.1091/mbc.E15-07-0483.
- Kotb, A.; Hyndman, M.E.; Patel, T.R. The role of zyxin in regulation of malignancies. Heliyon 2018, 4, e00695, doi:10.1016/j.heliyon.2018.e00695.
- Buday, L.; Downward, J. Roles of cortactin in tumor pathogenesis. Biophys. Acta BBA Rev. Cancer 2007, 1775, 263–273, doi:10.1016/j.bbcan.2006.12.002.
- MacGrath, S.M.; Koleske, A.J. Cortactin in cell migration and cancer at a glance. Cell Sci. 2012, 125, 1621–1626, doi:10.1242/jcs.093781.
- Mader, C.C.; Oser, M.; Magalhaes, M.A.O.; Bravo-Cordero, J.J.; Condeelis, J.; Koleske, A.J.; Gil-Henn, H. An EGFR–Src–Arg–Cortactin Pathway Mediates Functional Maturation of Invadopodia and Breast Cancer Cell Invasion. Cancer Res. 2011, 71, 1730–1741, doi:10.1158/0008-5472.CAN-10-1432.
- Chen, Y.-C.; Baik, M.; Byers, J.T.; Chen, K.T.; French, S.W.; Díaz, B. Experimental supporting data on TKS5 and Cortactin expression and localization in human pancreatic cancer cells and tumors. Data Brief 2019, 22, 132–136, doi:10.1016/j.dib.2018.11.138.
- Thuault, S.; Mamelonet, C.; Salameh, J.; Ostacolo, K.; Chanez, B.; Salaün, D.; Baudelet, E.; Audebert, S.; Camoin, L.; Badache, A. A proximity-labeling proteomic approach to investigate invadopodia molecular landscape in breast cancer cells. Rep. 2020, 10, 1–14, doi:10.1038/s41598-020-63926-4.
This entry is adapted from the peer-reviewed paper 10.3390/ijms21218117