Cervical cancer (CC) continues to be one of the leading causes of death for women across the world. Although it has been determined that papillomavirus infection is one of the main causes of the etiology of the disease, genetic and epigenetic factors are also required for its progression. Among the epigenetic factors are included the long noncoding RNAs (lncRNAs), transcripts of more than 200 nucleotides (nt) that generally do not code for proteins and have been associated with diverse functions such as the regulation of transcription, translation, RNA metabolism, as well as stem cell maintenance and differentiation, cell autophagy and apoptosis. Recently, studies have begun to characterize the aberrant regulation of lncRNAs in CC cells and tissues, including Homeobox transcript antisense RNA (HOTAIR), H19, Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), Cervical Carcinoma High-Expressed 1 (CCHE1), Antisense noncoding RNA in the inhibitors of cyclin-dependent kinase 4 (ANRIL), Growth arrest special 5 (GAS5) and Plasmacytoma variant translocation 1 (PVT1). They have been associated with several disease-related processes such as cell growth, cell proliferation, cell survival, metastasis and invasion as well as therapeutic resistance, and are novel potential biomarkers for diagnosis and prognosis in CC.
- cervical cancer
- epigenetics
- long noncoding RNAs
- biological function
- tumorigenesis
1. Cervical Cancer
Cervical cancer (CC) is the fourth most frequent cancer in women globally. It is a preventable disease and also curable if detected early and adequately treated. Yet it remains one of the most common cancers and the leading cause of cancer-related death in women across the globe [1].
CC occurs at the epithelium of the uterine cervix, particularly at the squamocolumnar junction of the ectocervix and endocervix, a site of continuous metaplastic activity [2]. Nearly all CCs are caused by human papillomavirus (HPV) infections, which is the most common sexually transmitted infection worldwide [3].
The two major histological types of epithelial tumors in the cervix are squamous cell carcinoma (SCC) and adenocarcinoma, the first being the most common one, comprising 95% of all diagnosed CCs worldwide, while the remaining 5% is represented by adenocarcinoma and other less common epithelial tumors [2].
The most common therapies used for CC treatment are radiotherapy and chemotherapy, but these methods are not always effective and can cause severe side effects. Hence, there is an evident interest in finding new biomarkers and novel treatment targets that could complement standard evaluation to determine the presence of cancer cells in cervical tissues [4,5].
Integration of high-risk HPV (HR-HPV) viral DNA into the host chromosomal DNA is the main pathological event involved in CC. HPV DNA is the only molecular marker developed for CC diagnosis. For this reason, it is necessary to evaluate other events that could be used as possible markers based on their clinical utility, such as chromosomal anomalies, cell-cycle checkpoints, DNA mutations, epigenetic regulation, among others [5].
There are some markers, such as P16, P16INK4A, P53, RB, E-cadherin, and Ki67, that Z showed the ability to be able to detect intraepithelial lesions that can evolve to invasive forms. Additionally, markers such as CEA, SCC-Ag and CD44 can detect invasive forms of the disease. Overall, several studies have been focused on detecting biomarkers capable of identifying molecular changes that lead to the development and progress of CC [6].
2. Noncoding RNAs in Cervical Cancer
Many noncoding RNAs (ncRNAs) were identified as molecular regulatory factors in cancer and may provide therapeutic targets for improving survival in cases of CC [4]. In recent years, it has been estimated that around 99% of the total RNA present in mammalian cells is comprised of ncRNA. Currently, it is not possible to know the exact amount of truly functional ncRNAs, as the number of these transcripts grows annually [7].
Based on their functions, the ncRNAs can be divided into two broad classes: housekeeping ncRNAs and regulatory ncRNAs. Housekeeping ncRNAs mainly regulate generic cellular functions—for instance, messenger RNA (mRNA) translation, splicing, and rRNA modification. On the other hand, regulatory ncRNAs can be divided, based on the length of the transcript, into short noncoding transcripts comprising less than 200 nucleotides (nt) and long noncoding RNAs comprising transcripts greater than 200 nt. [8]. This class of non-protein-coding RNA and functional RNA molecules include long noncoding RNAs (lncRNAs), microRNAs (miRNAs), small interfering RNA (siRNA), small nucleolar RNAs (snoRNA), piwi-interacting RNA (piRNA), among others [9,10].
The regulatory ncRNAs play critical roles in important biological processes such as DNA transcription, and mRNA post-transcriptional processing and translation. Furthermore, ncRNA can be packaged into exosomes and other extracellular vesicles, providing a mechanism for intercellular communication. Overwhelming evidence suggests that various ncRNAs can be implicated in human disease processes [9,10].
MiRNAs can target, by base-pairing, multiple mRNAs and modulate their expression. Many studies have reported the association of miRNAs and CC progression, involving mechanisms that affect apoptosis, cell proliferation, migration and invasion. Dysregulation of miRNAs at different stages of CC can play an important role in the development of the disease, as they are functionally involved in cell-cycle, P53, and Wnt signaling pathways, among others. Thus, stage-specific miRNAs could be used as CC biomarkers [11].
In addition to the role of the miRNAs in CC, other molecules have gained interest in oncology, since they are dysregulated in this pathology, including the lncRNAs.
2. LncRNAs Dysregulated in Cancer Cervical
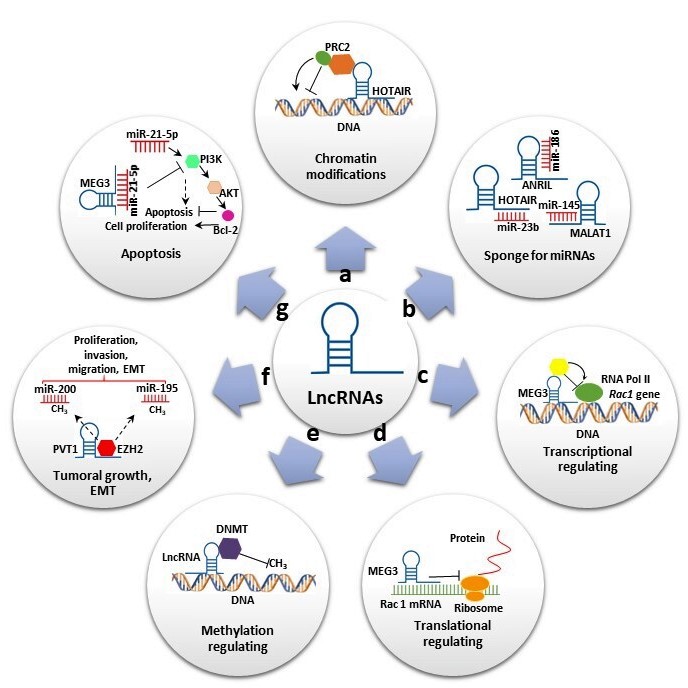
LncRNAs, defined as transcripts of more than 200 nt that generally do not code for proteins, have been associated with diverse functions and represent the largest class of ncRNAs. In contrast to short ncRNAs, which are mostly attributed to gene regulation, the mechanistic role of lncRNAs is highly diverse, increasing their functional complexity [6]. LncRNAs have been classified according to their relative location into sense lncRNAs, antisense lncRNAs, bidirectional lncRNAs, intron lncRNAs, intergenic lncRNAs and enhancer lncRNAs [12]. They participate in several biological processes at transcriptional, post-transcriptional and epigenetic levels (Figure 1). At the transcriptional level, lncRNAs induce or suppress gene expression by inducing chromatin modifications and or interacting with RNA polymerase, acting as miRNA sponges (Figure 1a,c). At the post-transcriptional level, lncRNAs can act as sponge for miRNAs (Figure 1b) and also can form a stable structure of double-stranded RNA with mRNAs and regulate the translation (Figure 1d). Additionally, they can bind to proteins and regulate their stability (Figure 1d). At the epigenetic level, the lncRNAs are involved in various mechanisms, including gene silencing throughDNAmethylation and reconstruction of chromatin conformation by acetylation, methylation or ubiquination of histones. (Figure 1e) [13]. In addition, some of these transcripts have been shown to play important roles in stem cell maintenance and di erentiation, cell autophagy, apoptosis and embryonic development (Figure 1f,g). Furthermore, lncRNAs were also reported as having oncogenic functions in many types of cancers and neurological and cardiovascular diseases [14].
Figure 1. Long noncoding RNAs’ (lncRNAs) biological roles. (a) Some lncRNAs regulate gene expression by assembling chromatin-modifying complexes. Homeobox transcript antisense RNA (HOTAIR) interacts with polycomb repressive complex 2 (PRC2) and establishes the repressive H3K27me3 chromatin mark. (b) lncRNAs can act as miRNA sponges. HOTAIR, Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) and Antisense noncoding RNA in the inhibitors of cyclin-dependent kinase 4 (ANRIL) sponge miR-23-b, miR-154 and miR-186, respectively, to reverse suppression of their target genes. (c,d) Some lncRNAs promote or suppress gene expression. Maternally expressed gene 3 (MEG3) inhibits Rac1 at both transcriptional and translational levels. (e) Some lncRNAs interact with diverse DNMT members, promoting or repressing DNA methylation. (f) Some lncRNAs play important roles in cell migration, proliferation and invasion. Plasmacytoma variant translocation 1 (PVT1) binds to EZH2, increasing H3K27me3 level on miR-195 and miR-200b promoters and promoting epithelial–mesenchymal transition (EMT). (g) lncRNAs also play significant roles in apoptosis. MEG3 reduce the level of miR-21-5p expression, inhibiting cell proliferation and increasing apoptosis by regulating the PI3K/AKT/BCL-2/Bax/P21 pathway.
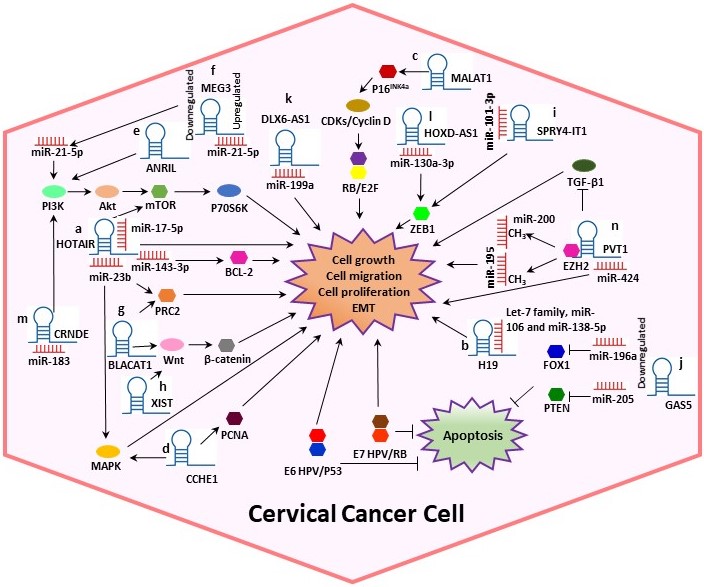
In CC, various studies have reported that lncRNAs are directly bound to target proteins or mRNAs to perform post-transcriptional modifications. Moreover, it has been suggested that lncRNAs play significant roles in CC progression by sponging miRNAs and interacting with HPV proteins [4,12]. In this context, we systematically summarized the dysregulation and the potential mechanisms of several lncRNAs such as Homeobox transcript antisense RNA (HOTAIR), H19, Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), Cervical Carcinoma High-Expressed 1 (CCHE1), Antisense noncoding RNA in the inhibitors of cyclin-dependent kinase 4 (ANRIL), Growth arrest special 5 (GAS5) and Plasmacytoma variant translocation 1 (PVT1), among others (Table 1, Figure 2), in cervical tumorigenesis.
Figure 2. Potential mechanisms of several lncRNAs in CC. Some lncRNAs play significant roles in CC progression by sponging miRNAs and interacting with other proteins, altering the cell cycle. (a) HOTAIR could act as a sponge for miR-143-3p, promoting BCL-2 expression. Also, HOTAIR exerts its tumor-promoting effect by sponging miR-17-5p and may indirectly modulate MAPK1 expression by binding to miR-23b. (b) H19 interacts with miRNAs of the let-7 family regulating diverse cellular processes. (c) MALAT1 may regulate cell proliferation through the P16INK4A/CDKs/RB pathway. (d) Cervical Carcinoma High-Expressed 1 (CCHE1) enhancing the expression of PCNA, CCHE1 also participates in the ERK/MAPK pathway. (e) ANRIL inhibition guides the inactivation of the PI3K/Akt pathway. (f) Maternally expressed gene 3 (MEG3) functions as a tumor-suppressor via regulating miR-21-5p. (g) Bladder cancer-associated transcript 1 (BLACAT1) binds to PRC2 and promotes proliferation, migration and invasion by modulating Wnt/β-catenin pathway. (h) X-inactive specific transcript (XIST) can play the role of an oncogene activating the Wnt/β-catenin pathway. (i) SPRY4-Intronic transcript or Sprouty4-Intronic transcript 1 (SPRY4-IT1) could bind to miR-101-3p to regulate the expression of the target gene ZEB1. (j) GAS5 expression is decreased in CC, leading to dysregulation of miR-196a and miR-205, which function as oncogenic miRNAs by targeting FOXO1 and PTEN, respectively. (k) DLX6 antisense RNA 1 (DLX6-AS1) may promote cell proliferation by sponging miR‑199a. (l) HOXD-AS1 upregulates ZEB1 through binding to miR-130a-3p. (m) CRNDE acts as an oncogene in CC through sponging miR-183. (n) PVT1 acts as a sponge for miR-424, miR-195 and miR-200b. Overall, the figure shows some summarized molecular mechanisms by which the lncRNAs promote cell growth, migration and proliferation and EMT in a CC cell.
Table 1. lncRNAs and their mechanisms and biological roles in Cervical cancer (CC).
|
LncRNA |
Localization |
Expression Level |
Signaling Pathways and Molecules |
Biological Roles |
Ref. |
|
HOTAIR |
12q13.13 |
Upregulated |
Notch/Wnt pathway, sponge for miR-143-3p, miR-23b and miR-17-5p. MAPK1, BCL-2, mTOR/p70S6K pathway. |
Cell growth, cell proliferation and cell survival, EMT metastasis, and invasion |
|
|
H19 |
11p15.5 |
Upregulated |
Sponge for let-7 family, miR-106, miR-194, miR-138-5p, miR-675, miR-140, miR-200 and MILK |
Cell migration, cell proliferation |
|
|
MALAT1 |
11q13 |
Upregulated |
P16INK4A/CDKs/RB pathway, sponge for miR-145. |
Cell migration, cell proliferation, vascular invasion, EMT, metastasis. |
|
|
CCHE1 |
10q21.1 |
Upregulated |
ERK/MAPK pathway. PCNA. |
Cell proliferation, metastasis. |
|
|
ANRIL |
9p21 |
Upregulated |
PI3K/Akt pathway, Sponge for miR-186. |
Cell proliferation, metastasis |
|
|
CCAT2 |
8q24.21 |
Upregulated |
Cell cycle |
Cell proliferation, metastasis. |
|
|
MEG3 |
14q32.3 |
Downregulated |
Sponge for miR-21-5p. PI3K/AKT/BCL-2/Bax/P21 pathway, PI3K/AKT/MMP-2/9 pathway, P-STAT3. |
Metastasis, apoptosis. |
|
|
BLACAT1 |
1q32.1 |
Upregulated |
Wnt/β-catenin pathway, sponge for miR-143 and miR-424. |
Cell proliferation, cell migration. |
|
|
XIST |
Xq13.2 |
Upregulated |
Wnt/β-catenin pathway, sponge for miR-140-5p. |
Cell proliferation, cell invasion, cell EMT, migration. |
|
|
SPRY4-IT1 |
5q31.3 |
Upregulated |
Sponge for miR-101-3p. ZEB1. |
Cell proliferation, cell migration |
|
|
GAS5 |
1q25.1 |
Downregulated |
Sponge for miR-196a and miR-205. |
Cell proliferation, cell invasion, apoptosis |
|
|
DLX6-AS1 |
7q21.3 |
Upregulated |
Sponge for miR-16-5p and miR-199a. ARPP19, FUS. |
Cell proliferation, cell invasion. |
|
|
HOXD-AS1 |
2q31.1 |
Upregulated |
Ras/ERK pathway, sponge for miR-130a-3p. ZEB1. |
Cell growth, cell migration, cell invasion. |
|
|
CRNDE |
16q12.2 |
Upregulated |
PI3K/AKT pathway, sponge for miR-183. CCNB1, PUMA, P53. |
Cell proliferation, apoptosis. |
|
|
PVT1 |
8q24.21 |
Upregulated |
Smad3, sponge for miR-140-5p, miR-424, miR-195 and miR-200b. EZH2, TGF-β1. |
Cell growth, cell proliferation, metastasis. |
The current studies and research on the functions and levels of the lncRNAs in CC open up new possibilities for the evaluation of molecular markers for the disease. However, even though the ability of lncRNAs to sponge specific miRNAs and to deregulate metabolic pathways has been investigated in cell culture models, tissues and CC cells, the biggest challenge regarding working with lncRNAs is the comprehension of the molecular mechanisms underlying their functions as well as their correlation with others ncRNAs (such as mRNAs, miRNAs, and circRNAs) and proteins, which is still not fully resolved. In the same way, lncRNAs’ interactions with the immune system, microenvironment, microbiome, hormonal milieu, or metabolome is not completely elucidated either.
Overall, we mention and briefly summarized some lncRNA that have great potential to be applied as biomarkers for the prognosis and metastasis in CC which currently remains a public health issue and one of the leading causes of women’s death worldwide. Studies concerning the association of lncRNAs in CC are still in their early stages, but recent evidence points towards the possible use of these ncRNAs as key molecular tools in understanding the disease. Therefore, more experimental and clinical studies are necessary for the exploration and characterization of the mechanisms by which the lncRNAs are involved in the prognosis, invasion, metastasis, chemoresistance and radioresistance in CC, offering novel alternative approaches for better diagnosis and therapy in the future.
This entry is adapted from the peer-reviewed paper 10.3390/ijms21249742