Microbes that cause infections amongst insects leading to death or serious disabilities are known as entomopathogens. Entomopathogenic bacteria and fungi are quite frequently found in soils and insect cadavers. The first step in utilizing these microbes as biopesticides is to isolate them, and several culture media and insect baiting procedures have been tested in this direction. In this work, the authors review the current techniques that have been developed so far, in the last five decades, and display brief protocols which can be adopted for the isolations of these entomopathogens. Among bacteria, this study focuses on Serratia spp. and bacteria from the class Bacilli. Among fungi, the review focuses those from the order Hypocreales, for example, genera Beauveria, Clonostachys, Lecanicillium, Metarhizium, and Purpureocillium. The authors chose these groups of entomopathogenic bacteria and fungi based on their importance in the microbial biopesticide market.
- Beauveria,Metarhizium,Hypocreales,Bacillus thuringiensis,Serratia
1. Isolation of Entomopathogenic Bacteria
Entomopathogenic bacteria are commonly found in soils. Hence, isolating insect-pathogenic strains is quite important. Different bacterial groups, such as symbionts of entomopathogenic nematode (EPN) Heterorhabditis spp. and Steinernema spp., i.e., Photorhabdus spp. and Xenorhabdus spp., and others, such as Yersinia entomophaga, Pseudomonas entomophila, and Chromobacterium spp., exhibit entomopathogenicity [18].
Entomopathogenic nematode symbiotic bacteria are isolated by dropping an insect’s hemolymph onto a nutrient bromothymol blue (0.0025% (w/v)) triphenyltetrazolium chloride (0.004% (w/v)) agar (NBTA) and incubating the streaked plate at 25 °C, and continuously subculturing until the uniform colonies are obtained [19]. Yersinia entomophaga is isolated by culturing the hemolymph of diseased larvae of New Zealand grass grub, Costelytra zealandica White (Coleoptera: Scarabaeidae), onto Luria-Bertani (LB) agar, followed by growth on Caprylate-thallous agar (CTA) (Appendix A, Medium 1) and Deoxyribonuclease (DNase)-Toluidine Blue agar (Appendix A, Medium 2), and no hemolysis on Columbia horse blood agar (Columbia agar + 5% horse blood) or Columbia sheep blood agar (Columbia agar + 5% sheep blood) [20]. Isolating P. entomophila is rather tricky as the bacterium needs to elicit the systemic expression of Diptericin, an antimicrobial peptide in Drosophila, after ingestion. However, the bacterial culture can be maintained on LB media [21]. Bacterial isolates from insects belonging to Chromobacterium exhibit violet pigment when cultured on L-agar [22]. However, EPB that are most commonly used as commercial biopesticides are further discussed in the review.
1.1. Milky Disease-Causing Paenibacillus spp.
Paenibacillus popilliae and Paenibacillus lentimorbus are obligate pathogens of scarabs (Coleoptera) as they require the host for the growth and sporulation. In soils, they are present as endospores. These bacteria can be isolated from the hemolymph, and the methodologies may vary depending on the bacterial species. The protocols listed below have been described by Stahly et al., and more details of these protocols have been reported by Koppenhöfer et al. [23,24,25].
- (a)
-
Disinfect the surface of the larvae of grubs (Coleoptera) with 0.5% (v/v) sodium hypochlorite (NaOCl).
- (b)
-
Pinch the cadaver using a sterilized needle and collect the emerging drops in sterilized water.
- (c)
-
Culture the dilutions of the drops on St. Julian medium (J-Medium) (Appendix A, Medium 1) [26], or Mueller-Hinton broth, yeast extract, potassium phosphate, glucose, and pyruvate (MYPGP) (Appendix A, Medium 2) agar [27].
Note: To enhance the germination of the vegetative cells, using 0.1% (w/v) tryptone solution is recommended during bacterial dilutions [26]. For spores, it is advisable to heat them for 15 min in a 1 M calcium chloride solution (pH 7.0) at 60 °C, and suspend them in the hemolymph of the cabbage looper Trichoplusia ni Hübner (Lepidoptera: Noctuidae) and in tyrosine at an alkaline pH. Another way to improve the germination is to heat the spores at 75 °C for 30 min and then apply pressure using a French press [28].
1.2. Amber Disease-Causing Serratia spp.
Serratia spp. are quite frequently isolated from soils, and some of them, being saprophytes, can also be isolated from insect cadavers. Therefore, to enhance the growth of insect pathogenic Serratia spp. such as Serratia entomophila, Serratia proteamaculans, and Serratia marcescens, a methodology based on a selective agar medium has been described by O’Callaghan and Jackson [30].
- (a)
-
Soil inoculums or hemolymph of the diseased larvae can be isolated on Caprylate-thallous agar (CTA) (Appendix A, Medium 3) [31].
- (b)
-
Culturing is done by pulling and separating the anterior end of the cadavers. The gut contents are then cultured on CTA plates.
- (c)
-
Serratia marcescens produces colonies that are red in color. Cream-colured bacterial colonies formed on CTA can then be transferred into different selective media for the identification of Serratia spp. [30].
- (d)
-
The production of a halo on a Deoxyribonuclease (DNase)-Toluidine Blue agar (Appendix A, Medium 4) when incubated at 30 °C for 24 h, indicates the presence of Serratia spp. [32]. Thereafter, the production of blue or green colonies on adonitol agar (Appendix A, Medium 5) confirms S. proteamaculans. The formation of yellow colonies on adonitol agar hints the presence of S. entomophila, which can be confirmed by the growth on itaconate agar (Appendix A, Medium 6) at 30 °C after 96 h [25]. Further molecular approaches targeting specific DNA regions can distinguish pathogenic strains from the non-pathogenic ones.
2. Isolation of Entomopathogenic Fungi
Fungal entomopathogens can directly be isolated from insect cadavers in the case of visible mycosis [35]. Moreover, they can also be isolated from soils or phylloplane as they spend a considerable part of their life as saprophytes in soils or as plant endophytes. However, to our knowledge, their survival as soil saprophytes has not been proven yet [4,5,6,7,8,35,36]. In either case, the material can be cultured directly onto a medium selective for an EPF or the material can be baited with an infection-sensitive insect [37]. In case of the isolation of EPF as endophyte, proper disinfection of the material is needed. Nonetheless, different antibacterial and fungal saprophyte-inhibiting chemicals are added in the selective medium, as per the research interest. Here, different culture media used to isolate fungal entomopathogens, especially those belonging to the order Hypocreales are discussed.
2.1. Isolations from Naturally Mycosed Insect Cadavers
This method is applied to study the natural EPF infections in the fields as it relies on the collection of the dead insects from the fields. The protocol described below is similar to that employed by Sharma et al. [7].
- (a)
-
Insect cadavers are brought to the laboratory as separate entities in sterile tubes.
- (b)
-
Insects are observed under a stereomicroscope (40×) for probable mycosis.
- (c)
-
In case of a visible mycosis, the insects are surface sterilized using 70% ethanol or 1% NaOCl, for 3 min, followed by 3 distinct washes with 100 mL of sterilized water. Then, the sporulating EPF from the insect cadaver is plated directly.
- (d)
-
Cadavers are then cultured on a selective medium at 22 °C for up to 3 weeks, depending on the time taken by the fungi for germination and proliferation. In case of no germination, the cadavers can be homogenized and plated on the selective medium. Details of the different selective medium are provided later in the text.
- (e)
-
Obtained fungi are subcultured on potato dextrose agar (PDA) (Appendix A, Medium 8) or Sabouraud dextrose agar (SDA) (Appendix A, Medium 9) until pure culture is obtained.
- (f)
- (g)
-
Molecular identifications can be done by extracting the DNA and performing PCR for the amplification and subsequent sequencing of the nuclear internal transcribed spacer (nrITS) region of the fungal nuclear ribosomal DNA, as described in Yurkov et al. [40].
2.2. Isolations from Soils
2.2.1. Soil Suspension Culture
This method is generally used to isolate a particular EPF genus of interest using different concentrations of the soil inoculums. To ensure correct isolation, the isolated EPF should also be characterized morphologically and molecularly, as described in Section 3.1. Here the authors discuss various selective media used, especially those which are useful for the isolation of the hypocrealean fungi pertaining to their dominance in fungi-based microbial pesticide market.
Metarhizium spp.
Isolating EPF has always been challenged by the contamination from saprophytic fungi. In this direction, Veen and Ferron [49] suggested using dodine (N-dodecylguanidine monoacetate) to inhibit the growth of saprophytes and developed Veen’s semi-selective medium to accomplish this (Appendix A, Medium 12). Later, Chase et al. [50] and Sneh [51] also used dodine in their studies. However, Liu et al. [52] reported that the higher quantities of dodine can be inhibitory to EPF and suggested using only 10 µg/mL dodine (Appendix A, Medium 12). Later, Rangel et al. [53] cautioned against the use of dodine and showed the even 0.006% (w/v) dodine in PDAY can completely inhibit Metarhizium acridum. This led to the development of CTC medium, which is made by the addition of 0.05% (w/v) chloramphenicol, 0.0001% (w/v) thiabendazole, and 0.025% (w/v) cycloheximide in PDAY [54] (Appendix A, Medium 13). However, a recent study by Hernández-Domínguez et al. [55] suggested the use of CTC medium, along with other dodine-containing mediums, for better Metarhizium recoveries. Posadas et al. [47] demonstrated that OM-CTAB is effective in isolating EPF while inhibiting saprophytes. Moreover, this negated the dependency on dodine, as it is not easily available in some countries.
Beauveria spp.
Beauveria spp., e.g., Beauveria bassiana sensu lato (s.l.) and Beauveria pseudobassiana, can be easily isolated using oatmeal dodine agar (ODA), as described by Chase et al. [50] (Appendix A, Medium 14). This medium has also been used in recent studies [56,57,58,59]. Another medium, i.e., Sabouraud-2-glucose agar (S2GA), was made by Strasser et al. [60] (Appendix A, Medium 15) for the isolation of Beauveria brongniartii, and was successfully used in studies concerning B. brongniartii [61,62,63]. However, many recent studies have used S2GA, with slight modifications, to isolate B. bassiana s.l. [64,65]. A dodine-free alternative in isolating B. bassiana s.l. is OM-CTAB [47]. Moreover, Ramírez-Rodríguez and Sánchez-Peña [66] suggested using PDAY with CTAB (0.015% or 0.03% (w/v)) and any of the antibacterial compounds, i.e., dihydrostreptomycin, oxytetracycline, or doxycycline, to isolate Beauveria while inhibiting fungal saprophytes.
Purpureocillium spp.
Purpureocillium spp., i.e., Purpureocillium lilacinum and Purpureocillium lavendulum, can easily be isolated using an agar medium containing sodium chloride, benomyl, pentachloronitrobenzene, and Tergitol [67,68] (Appendix A, Medium 16).
Lecanicillium spp.
A Lecanicillium-selective medium (LSM) was developed by Kope et al. [69]. OM agar with 0.05% (w/v) chloramphenicol and 0.05% (w/v) CTAB can also be used, as described recently by Xie et al. [70] (Appendix A, Medium 17).
Clonostachys spp.
Clonostachys spp., e.g., Clonostachys rosea f. rosea, is reported entomopathogenic and can be isolated frequently from soils. Culture medium such as DRBCA is highly effective in isolating Clonostachys spp., at least in the case of the isolations from cadavers [7].
2.2.2. Insect Baiting
This method is arguably the most commonly used method for EPF isolation, as the bait insect specifically selects entomopathogens from other saprobes in the soils [35,71,72], although surface sterilization of the insect cadavers is needed to avoid occasional contaminations by saprophytic fungi.
Galleria-Bait Method or Tenebrio-Bait Method
The use of Galleria mellonella Linnaeus (Lepidoptera: Pyralidae) for isolating EPF from soil or the “Galleria-bait method” was first described by Zimmermann [73]. Since then, it has been used for EPF isolations in many studies [74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91]. Tenebrio molitor Linnaeus (Coleoptera: Tenebrionidae) has also been used as a bait insect in some studies [92,93,94]. Some previous studies have noticed that insect baiting is more sensitive in isolating EPF than culturing soil suspensions on selective medium [61,62,95,96]. Other studies have also used insect baiting along with soil suspension cultures [57,97,98,99,100]. Although insect baiting is a widely accepted method for EPF isolation, it should be used with caution as some lines of insect baits, such as the dark (melanic) morphs of G. mellonella, are more resistant to B. bassiana s.l., and this trait has also been observed in T. molitor for M. anisopliae s.l. [101,102]. Similarly, immune-suppressed G. mellonella were found to be highly (~200 times) susceptible to EPF, which can lead to the isolation of a diverse set of EPF from soils, although saprophytic fungi may not induce any insect mortality [103].
Galleria-Tenebrio-Bait Method
As bait insects can be sensitive to infection by one particular EPF genus, some studies have used both G. mellonella and T. molitor to isolate EPF, either in part or throughout their whole experiment [7,104,105,106,107]. Recently, Sharma et al. [7] suggested using the “Galleria-Tenebrio-bait method” to avoid any underestimation of EPF abundance and diversity, as it was found that G. mellonella and T. molitor were significantly more sensitive toward the infections by B. bassiana s.l. and M. robertsii, respectively. This method is described in Figure 2.
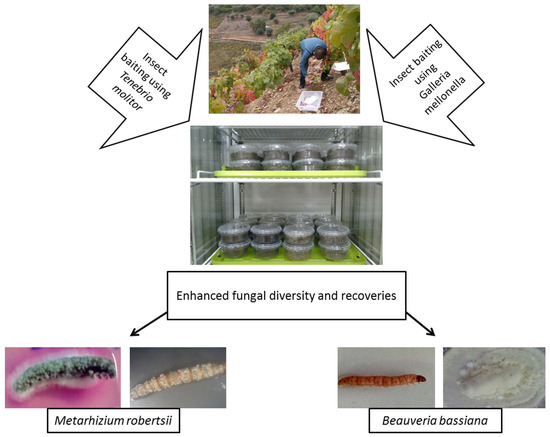
2.3. Isolation from Phyllosphere
Some studies have also isolated EPF from the phylloplane and other parts of the plant phyllosphere, as these fungi can also be present as plant epiphytes or endophytes [41]. Meyling et al. suggested a leaf imprinting methodology where the leaf is cultured onto a selective agar medium [64]. Petri dishes with partitions are used and the upper (adaxial), and the lower (abaxial) surface of the leaf are pressed on the separate sides of the Petri plate. Henceforth, the same leaf is put on a paper sheet and photocopied to estimate its surface area using image analysis software at a later stage. The petri plates are incubated in the dark at 23 °C to count fungal colony-forming units (CFUs) [64]. Surface sterilization is quite important in isolating hypocrealean fungi as endophytes. This can be done by dipping the plant part in either 70% ethanol and/or 1–5% NaOCl for 3 min. In the case of the leaves, the petiole can be first kept out of the sanitizer to avoid the chemical reaching inside the leaf, and then it can be cut to culture the sterilized part of the leaf on either of the selective mediums described above. It is always recommended to sanitize the intact plant part and then cut it into pieces for further culturing, as this avoids the sterilization of the endophytic fungi [111]. Different studies have isolated EPF from the phyllosphere, such as bark and branch samples [56,112] and leaves [59,113].
2.4. Molecular Identifications of the Isolated Entomopathogenic Fungi
After obtaining a single spore fungal culture on a PDA or SDA (Appendix A; Medium 8 and/or 9), as described in the Section 3.1, the species can be resolved or identified by amplifying the regions of nuclear ribosomal DNA, such as nrITS, large (28S) subunit (nrLSU), or small (18S) subunit (nrSSU). Another, nuclear ribosomal DNA region, i.e., the intergenic spacer region between nrSSU and nrLSU or IGS, has also been used to understand Beauveria and Metarhizium speciation [113,114,115,116]. The resolution of the molecular identification can be increased by amplifying other nuclear DNA regions of interest, e.g., for Bloc for Beauveria [113,114,115] and the 5′ intron-containing region of translation elongation factor 1-alpha subunit (5′-tef1α) for Metarhizium [116,117]. Other nuclear DNA markers, such as the regions of the gene encoding for the largest subunit of RNA polymerase II (rpb1), the second largest subunit of RNA polymerase II (rpb2); β-tublin (β-tub), and the coding region of Tef1-α, can also be employed, in general, for any EPF [118,119].
Moreover, in the last decades, researchers have been constantly developing and validating the use of several microsatellite markers for the genotyping of Beauveria [93,115,120,121,122,123] and Metarhizium [124,125] isolates. For example, Oulevey et al. [125] described 18 small single repeats or microsatellite marker sets for Metarhizium, i.e., Ma145, Ma325, Ma307, Ma2049, Ma2054, Ma2055, Ma2056, Ma2057, Ma2060, Ma2063, Ma2069, Ma2070, Ma2077, Ma2089, Ma2283, Ma2287, Ma2292, and Ma2296. Similarly, Meyling et al. [93] and Goble et al. [123] validated the use of 17 to 18 microsatellite marker sets for Beauveria, i.e., Ba06, Ba08, and Ba12-Ba29. This methodology enables enhanced resolution among very closely related isolates which may otherwise be rendered as clones. Recently, Kepler and Rehner [119] developed primers for the amplification and sequencing of nuclear intergenic spacer markers for the resolution of Metarhizium isolates, i.e., BTIGS, MzFG543, MzFG546, MzIGS2, MzIGS3, MzIGS5, and MzIGS7, and Kepler et al. [99] successfully validated the use of MzIGS3 and MzFG543 on the Metarhizium isolated from agricultural soils.
This entry is adapted from the peer-reviewed paper 10.3390/microorganisms9010016
