KMT2 (histone-lysine N-methyltransferase subclass 2) complexes methylate lysine 4 on the histone H3 tail at gene promoters and gene enhancers. H3K4 methylation mark allows to control gene transcription. The KMT2s function in large multi-subunit complexes, which, in vertebrates, are often referred to as COMPASS or COMPASS-like complexes (COMplex of Proteins ASsociated with Set1). These complexes contain an enzyme (KMT2A or KMT2B, KMT2C or KMT2D, KMT2F or KMT2G), common core subunits (WDR5, RBBP5, ASH2L, DPY30) and unique interacting proteins, which are different for each of the three KMT2 groups (A/B, C/D and F/G). Also, the KMT2 complexes dynamically interact with many transcription factors.
- histone-lysine N-methyltransferase subclass 2
- gene transcription
- normal development
- aberrant growth
1. Introduction
KMT2 (histone-lysine N-methyltransferase subclass 2) complexes methylate lysine 4 on the histone H3 tail at gene promoters and gene enhancers and, thus, control the process of gene transcription. These complexes not only play an essential role in normal development but have also been described as involved in the aberrant growth of tissues.
2. Structure of the KMT2 Complexes
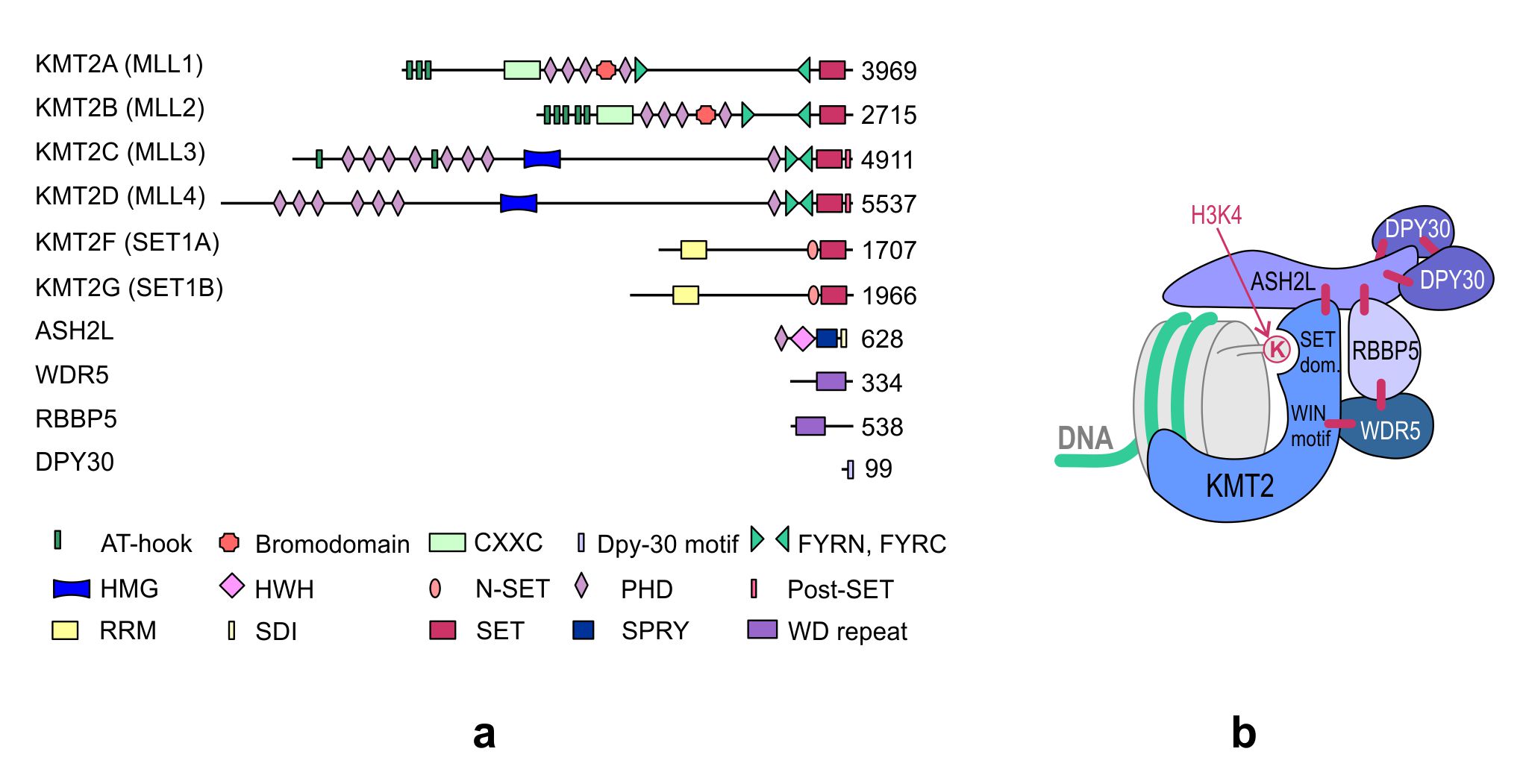
KMT2s are the main H3K4 methyltransferases that regulate gene transcription. The KMT2 family is highly conserved in eukaryotes. While three subgroups of KMT2s, each with a single representative, are present in Drosophila melanogaster—trithorax (Trx), trithorax-related (Trr), and Set1—two paralogs of each subgroup appeared in humans during evolution. Human cells contain two Trx-related KMT2s (KMT2A and KMT2B), two Trr-related KMT2s (KMT2C and KMT2D), and two Set1-related KMT2s (KMT2F and KMT2G) (Figure 1). Although KMT2E was initially classified under the KMT2 family, it was identified to be more homologous to yeast SET3 (SET domain-containing protein 3) and SET4 (SET domain-containing protein 4) and Drosophila CG9007 (encoding the protein “UpSET”) [1][2][3]. Moreover, unlike the other KMT2s present in humans, KMT2E does not exhibit intrinsic methyltransferase activity toward histone substrates [3].
Figure 1. (a) Domain structure of the KMT2 family and core subunits of the KMT2 complexes. The numbers indicate the number of amino acids. KMT, histone–lysine N-methyltransferase; ASH2L, absent, small, or homeotic 2-like; DPY30, Dumpy-30; RBBP5, retinoblastoma-binding protein 5; WDR5, WD repeat-containing protein 5; AT-hook, adenosine-thymidine-hook; CXXC, Zinc finger-CXXC domain; FYRN/FYRC, phenylalanine and tyrosine-rich region (N- and C-terminal); HMG, high mobility group; HWH, helix-wing-helix domain; N-SET, N-terminal of SET; PHD, plant homeodomain; Post-SET, C-terminal of SET; RRM, RNA recognition motif; SDI, Sdc1-Dpy-30 interaction; SET, Su(var)3-9, Enhancer-of-zeste and Trithorax; SPRY, SPla and the ryanodine receptor domain; and WD repeat, tryptophan-aspartic acid repeat. (b) The structure of the KMT2 complex. The enzyme and the core subunits of the complex are shown in the diagram. The interactions between individual subunits are marked with blue lines. Subunits specific to individual KMT2 complexes, not shown in the figure, interact with the amino terminus of the KMT2s. WIN motif, WDR5 interaction motif.
The KMT2s function in large multi-subunit complexes, which, in vertebrates, are often referred to as COMPASS or COMPASS-like complexes (COMplex of Proteins ASsociated with Set1). While there is one COMPASS complex in yeast, there are three in Drosophila and six closely related complexes in vertebrates, which contain one KMT2 methyltransferase unique to each complex, four core subunits commonly found in all KMT2 complexes, and additional complex-specific proteins (Table 1).
Table 1. Subunit composition of the mammalian KMT2 complexes.
|
|
KMT2A or KMT2B Complex |
KMT2C or KMT2D Complex |
KMT2F or KMT2G Complex |
|
Enzyme |
KMT2A or KMT2B |
KMT2C or KMT2D |
KMT2F or KMT2G |
|
Core subunits |
ASH2L RBBP5 WDR5 DPY30 |
ASH2L RBBP5 WDR5 DPY30 |
ASH2L RBBP5 WDR5 DPY30 |
|
Unique subunits |
Menin HCF1 or HCF2 |
PTIP PA1 NCOA6 UTX |
CFP1 WDR82 HCF1 |
KMT, histone–lysine N-methyltransferase; ASH2L, absent, small, or homeotic 2-like; RBBP5, retinoblastoma-binding protein 5; WDR5, WD repeat-containing protein 5; DPY30, Dumpy-30; HCF1, host cell factor 1; PTIP, PAX transactivation-domain interacting protein; PA1, PTIP-associated 1; NCOA6, nuclear receptor coactivator 6; UTX, ubiquitously transcribed tetratricopeptide repeat, X chromosome; CFP1, CXXC finger protein 1; and WDR82, WD repeat-containing protein 82.
The four core subunits—WDR5 (WD repeat domain 5), RBBP5 (retinoblastoma-binding protein 5), ASH2L (absent, small or homeotic 2-like), and DPY30 (Dumpy-30)—form a subcomplex that stably interacts with the KMT2 enzymes and stimulates KMT2 catalytic activity up to several hundred-fold [4][5]. A crystal structure analysis of the KMT2A complex has shown that the interaction of KMT2A with the core subunits (WDR5, RBBP5, and ASH2L) forces a conformational change in the SET domain of KMT2A, which is necessary to achieve a catalytically efficient form [5][6][7][8]. Biochemical and structural studies have demonstrated that the KMT2A complex is stabilized by direct interactions between KMT2A and WDR5 that bridges KMT2A to RBBP5. Furthermore, RBBP5 interacts with ASH2L, which binds the DPY30 protein. Although WDR5 is essential for regulating the activity of KMT2A, it is not responsible for the regulation of other KMT2s, and these KMT2s have been shown to be stimulated by the stable ASH2L-RBBP5 heterodimer that directly interacts with them [4]. The structure of the KMT2 complex is presented in Figure 1b.
In addition to their role in stabilizing and regulating KMT2 complexes, the core subunits of the WRAD subcomplex participate in the recruitment of these complexes to chromatin. The recruitment of KMT2 complexes to the genomic loci is also regulated by the unique complex-specific subunits that determine the functional diversity of these complexes. The three KMT2 groups of complexes differ in their sets of interacting proteins (Table 1). The additional subunits associated with the KMT2A/KMT2B complexes are Menin and HCF1/2 (host cell factors 1/2) [9][10]. Another protein that interacts with the KMT2A/KMT2B complexes is LEDGF (lens epithelium-derived growth factor, also known as PSIP1/p75). However, LEDGF is a substoichiometric component in these complexes and exhibits only a weak interaction with them through Menin [10][11][12]. PTIP (PAX transactivation domain-interacting protein), PA1 (PTIP-associated 1), NCOA6 (nuclear receptor coactivator 6), and UTX (ubiquitously transcribed tetratricopeptide repeat, X chromosome) interact specifically with KMT2C and KMT2D [13][14][15], while CFP1 (CXXC finger protein 1), WDR82 (WD repeat domain 82), and HCF1 (host cell factor 1) interact only with KMT2G and KMT2F [10][16].
In addition to the stable interaction with the complex-specific subunits, the KMT2 complexes dynamically interact with many transcription factors, including NFE2 (nuclear factor, erythroid 2) [17], USF1 (upstream transcription factor 1) [18], MEF2D (myocyte enhancer factor 2D) [19], NF-Y (nuclear transcription factor Y) [20], NF-E2 (nuclear transcription factor erythroid 2) [17][21], AP2δ (activating protein 2δ) [22], MYC [23], OCT4 (octamer-binding transcription factor 4, also known as POU5F1) [24], NANOG [25], p53 [26][27][28], E2Fs [26][29][30], and PAX7 (Paired Box 7) [31]. These interactions of the KMT2 complexes are important for their context-dependent roles.
This entry is adapted from the peer-reviewed paper 10.3390/ijms21249340
References
- Joel C. Eissenberg; Ali Shilatifard; Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Developmental Biology 2010, 339, 240-249, 10.1016/j.ydbio.2009.08.017.
- W.W.M. Pim Pijnappel; Daniel Schaft; Assen Roguev; Anna Shevchenko; Hille Tekotte; Matthias Wilm; Guillaume Rigaut; Bertrand Séraphin; Rein Aasland; A. Francis Stewart; et al. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes & Development 2001, 15, 2991-3004, 10.1101/gad.207401.
- XiaoMing Zhang; Wisna Novera; Yan Zhang; Lih-Wen Deng; MLL5 (KMT2E): structure, function, and clinical relevance. Cellular and Molecular Life Sciences 2017, 74, 2333-2344, 10.1007/s00018-017-2470-8.
- Yali Dou; Thomas A Milne; Alexander J Ruthenburg; Seunghee Lee; Jae Woon Lee; Gregory L Verdine; C David Allis; Robert G Roeder; Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nature Structural & Molecular Biology 2006, 13, 713-719, 10.1038/nsmb1128.
- Anamika Patel; Venkatasubramanian Dharmarajan; Valarie E. Vought; Michael S. Cosgrove; On the Mechanism of Multiple Lysine Methylation by the Human Mixed Lineage Leukemia Protein-1 (MLL1) Core Complex. Journal of Biological Chemistry 2009, 284, 24242-24256, 10.1074/jbc.m109.014498.
- Vanja Avdic; Pamela Zhang; Sylvain Lanouette; Adam Groulx; Véronique Tremblay; Joseph Brunzelle; Jean-François Couture; Structural and Biochemical Insights into MLL1 Core Complex Assembly. Structure 2011, 19, 101-108, 10.1016/j.str.2010.09.022.
- Jean-François Couture; Georgios Skiniotis; Assembling a COMPASS. Epigenetics 2013, 8, 349-354, 10.4161/epi.24177.
- Stacey M. Southall; Poon-Sheng Wong; Zain Odho; S. Mark Roe; Jon R. Wilson; Structural Basis for the Requirement of Additional Factors for MLL1 SET Domain Activity and Recognition of Epigenetic Marks. Molecular Cell 2009, 33, 181-191, 10.1016/j.molcel.2008.12.029.
- Christina M. Hughes; Orit Rozenblatt-Rosen; Thomas A. Milne; Terry D. Copeland; Stuart S. Levine; Jeffrey C. Lee; D. Neil Hayes; Kalai Selvi Shanmugam; Arindam Bhattacharjee; Christine A. Biondi; et al. Menin Associates with a Trithorax Family Histone Methyltransferase Complex and with the Hoxc8 Locus. Molecular Cell 2004, 13, 587-597, 10.1016/s1097-2765(04)00081-4.
- Rick Van Nuland; Arne H. Smits; Paschalina Pallaki; Pascal W. T. C. Jansen; Michiel Vermeulen; H. Th. M. Timmers; Quantitative Dissection and Stoichiometry Determination of the Human SET1/MLL Histone Methyltransferase Complexes. Molecular and Cellular Biology 2013, 33, 2067-2077, 10.1128/MCB.01742-12.
- Marcelo J. Murai; Jonathan Pollock; Shihan He; Hongzhi Miao; Trupta Purohit; Adam Yokom; Jay L. Hess; Andrew G. Muntean; Jolanta Grembecka; Tomasz Cierpicki; et al. The same site on the integrase-binding domain of lens epithelium–derived growth factor is a therapeutic target for MLL leukemia and HIV. Blood 2014, 124, 3730-3737, 10.1182/blood-2014-01-550079.
- Akihiko Yokoyama; Michael L. Cleary; Menin Critically Links MLL Proteins with LEDGF on Cancer-Associated Target Genes. Cancer Cell 2008, 14, 36-46, 10.1016/j.ccr.2008.05.003.
- Young-Wook Cho; Teresa Hong; SunHwa Hong; Hong Guo; Hong Yu; Doyeob Kim; Tad Guszczynski; Gregory R. Dressler; Terry D. Copeland; Markus Kalkum; et al. PTIP Associates with MLL3- and MLL4-containing Histone H3 Lysine 4 Methyltransferase Complex. Journal of Biological Chemistry 2007, 282, 20395-20406, 10.1074/jbc.m701574200.
- Young-Hwa Goo; Young Chang Sohn; Dae-Hwan Kim; Seung-Whan Kim; Min-Jung Kang; Dong-Ju Jung; Eunyee Kwak; Nickolai A. Barlev; Shelley L. Berger; Vincent T. Chow; et al. Activating Signal Cointegrator 2 Belongs to a Novel Steady-State Complex That Contains a Subset of Trithorax Group Proteins. Molecular and Cellular Biology 2003, 23, 140-149, 10.1128/mcb.23.1.140-149.2003.
- Sanjeevkumar R. Patel; Doyeob Kim; Inna Levitan; Gregory R. Dressler; The BRCT-Domain Containing Protein PTIP Links PAX2 to a Histone H3, Lysine 4 Methyltransferase Complex. Developmental Cell 2007, 13, 580-592, 10.1016/j.devcel.2007.09.004.
- Jeong-Heon Lee; Courtney M. Tate; Jin-Sam You; David G. Skalnik; Identification and Characterization of the Human Set1B Histone H3-Lys4Methyltransferase Complex. Journal of Biological Chemistry 2007, 282, 13419-13428, 10.1074/jbc.m609809200.
- Celina Demers; Chandra-Prakash Chaturvedi; Jeffrey A. Ranish; Gaetan Juban; Patrick Lai; Francois Morle; Ruedi Aebersold; F. Jeffrey Dilworth; Mark Groudine; Marjorie Brand; et al. Activator-Mediated Recruitment of the MLL2 Methyltransferase Complex to the β-Globin Locus. Molecular Cell 2007, 27, 573-584, 10.1016/j.molcel.2007.06.022.
- Changwang Deng; Ying Li; Shermi Liang; Kairong Cui; Tal Salz; Hui Yang; Zhanyun Tang; Patrick G. Gallagher; Yi Qiu; Robert Roeder; et al. USF1 and hSET1A Mediated Epigenetic Modifications Regulate Lineage Differentiation and HoxB4 Transcription. PLOS Genetics 2013, 9, e1003524, 10.1371/journal.pgen.1003524.
- Arif Aziz; Qi-Cai Liu; F. Jeffrey Dilworth; Regulating a master regulator. Epigenetics 2010, 5, 691-695, 10.4161/epi.5.8.13045.
- Andrea Fossati; Diletta Dolfini; Giacomo Donati; Roberto Mantovani; NF-Y Recruits Ash2L to Impart H3K4 Trimethylation on CCAAT Promoters. PLOS ONE 2011, 6, e17220, 10.1371/journal.pone.0017220.
- Aeri Kim; Sang-Hyun Song; Marjorie Brand; Ann Dean; Nucleosome and transcription activator antagonism at human β-globin locus control region DNase I hypersensitive sites. Nucleic Acids Research 2007, 35, 5831-5838, 10.1093/nar/gkm620.
- Cheryl C. Tan; K. V. Sindhu; Side Li; Hitomi Nishio; Jason Z. Stoller; Kimihiko Oishi; Sahitya Puttreddy; Tamara J. Lee; Jonathan A. Epstein; Martin J. Walsh; et al. Transcription factor Ap2 associates with Ash2l and ALR, a trithorax family histone methyltransferase, to activate Hoxc8 transcription. Proceedings of the National Academy of Sciences 2008, 105, 7472-7477, 10.1073/pnas.0711896105.
- Andrea Ullius; Juliane Lüscher-Firzlaff; Ivan G. Costa; Gesa Walsemann; Alexandra H. Forst; Eduardo G. Gusmao; Karsten Kapelle; Henning Kleine; Elisabeth Kremmer; Jörg Vervoorts; et al. The interaction of MYC with the trithorax protein ASH2L promotes gene transcription by regulating H3K27 modification.. Nucleic Acids Research 2014, 42, 6901-20, 10.1093/nar/gku312.
- Yen-Sin Ang; Su-Yi Tsai; Dung-Fang Lee; Jonathan Monk; Jie Su; Kajan Ratnakumar; Junjun Ding; Yongchao Ge; Henia Darr; Betty Chang; et al. Wdr5 Mediates Self-Renewal and Reprogramming via the Embryonic Stem Cell Core Transcriptional Network. Cell 2011, 145, 183-197, 10.1016/j.cell.2011.03.003.
- Alessandro Bertero; Pedro Madrigal; Antonella Galli; Nina C. Hubner; Inmaculada Moreno; Deborah Burks; Stephanie Brown; Roger A. Pedersen; Daniel Gaffney; Sasha Mendjan; et al. Activin/Nodal signaling and NANOG orchestrate human embryonic stem cell fate decisions by controlling the H3K4me3 chromatin mark. Genes & Development 2015, 29, 702-717, 10.1101/gad.255984.114.
- Yali Dou; Thomas A. Milne; Alan J. Tackett; Edwin R. Smith; Aya Fukuda; Joanna Wysocka; C. David Allis; Brian T. Chait; Jay L. Hess; Robert G. Roeder; et al. Physical Association and Coordinate Function of the H3 K4 Methyltransferase MLL1 and the H4 K16 Acetyltransferase MOF. Cell 2005, 121, 873-885, 10.1016/j.cell.2005.04.031.
- Jeongkyung Lee; Dae-Hwan Kim; Seunghee Lee; Qi-Heng Yang; Dong Kee Lee; Robert G. Roeder; A tumor suppressive coactivator complex of p53 containing ASC-2 and histone H3-lysine-4 methyltransferase MLL3 or its paralogue MLL4. Proceedings of the National Academy of Sciences 2009, 106, 8513-8518, 10.1073/pnas.0902873106.
- Zhanyun Tang; Wei-Yi Chen; Miho Shimada; Uyen T.T. Nguyen; Jaehoon Kim; Xiao-Jian Sun; Toru Sengoku; Robert K. McGinty; Joseph P. Fernandez; Tom W. Muir; et al. SET1 and p300 Act Synergistically, through Coupled Histone Modifications, in Transcriptional Activation by p53. Cell 2013, 154, 297-310, 10.1016/j.cell.2013.06.027.
- Shugaku Takeda; David Y Chen; Todd D. Westergard; Jill K. Fisher; Jeffrey A. Rubens; Satoru Sasagawa; Jason T. Kan; Stanley J. Korsmeyer; Emily H.-Y. Cheng; James J. Hsieh; et al. Proteolysis of MLL family proteins is essential for Taspase1-orchestrated cell cycle progression. Genes & Development 2006, 20, 2397-2409, 10.1101/gad.1449406.
- Shweta Tyagi; Anna Lena Chabes; Joanna Wysocka; Winship Herr; E2F Activation of S Phase Promoters via Association with HCF-1 and the MLL Family of Histone H3K4 Methyltransferases. Molecular Cell 2007, 27, 107-119, 10.1016/j.molcel.2007.05.030.
- Yoh-Ichi Kawabe; Yu Xin Wang; Iain W. McKinnell; Mark T. Bedford; Michael A. Rudnicki; Carm1 Regulates Pax7 Transcriptional Activity through MLL1/2 Recruitment during Asymmetric Satellite Stem Cell Divisions. Cell Stem Cell 2012, 11, 333-345, 10.1016/j.stem.2012.07.001.