Contamination by pesticides in the food chain and the environment is a worldwide problem that needs to be actively monitored to ensure safety. Unfortunately, standard pesticide analysis based on mass spectrometry takes a lot of time, money and effort. Thus, simple, reliable, cost-effective and field applicable methods for pesticide detection have been actively developed. One of the most promising technologies is an aptamer-based biosensor or so-called aptasensor. It utilizes aptamers, short single-stranded DNAs or RNAs, as pesticide recognition elements to integrate with various innovative biosensing technologies for specific and sensitive detection of pesticide residues. Several platforms for aptasensors have been dynamically established, such as colorimetry, fluorometry, electrochemistry, electrochemiluminescence (ECL) and so forth. Each platform has both advantages and disadvantages depending on the purpose of use and readiness of technology. For example, colorimetric-based aptasensors are more affordable than others because of the simplicity of fabrication and resource requirements. Electrochemical-based aptasensors have mainly shown better sensitivity than others with exceedingly low detection limits. This paper critically reviews the progression of pesticide aptasensors throughout the development process, including the selection, characterization and modification of aptamers, the conceptual frameworks of integrating aptamers and biosensors, the ASSURED (affordable, sensitive, specific, user-friendly, rapid and robust, equipment-free and deliverable to end users) criteria of different platforms and the future outlook.
- aptamers
- SELEX
- pesticides
- biosensors
- ASSURED
1. Generation of Aptamers against Pesticides
An aptamer requires either single-stranded DNA or RNA with a length of approximately 15–100 nucleotides to maintain its specific recognition and discrimination of the cognate target in a heterogeneous milieu [1]. Aptamers can also be found in the form of peptides [2]; Naturally, aptamers can be found in living organisms in the form of riboswitches, which function by binding with their ligands for subsequent regulation of specific gene expression [3]. Aptamers generated in vitro can be systematically isolated from a large pool of 1012–1015 synthetic oligo sequences using an iterative process called systematic evolution of ligands by exponential enrichment (SELEX). Since being first established in 1990, concurrently by Ellington and Szostak—who isolated dye-specific RNA [4]—and Tuerk and Gold—who developed T4 DNA polymerase-specific RNA [5]—the SELEX process has been applied and modified for the discovery of aptamers against a broad range of targets, such as proteins [6], metals [7], toxins [8], bacteria [9] and cancerous cells [10]. The details of the conventional SELEX procedure have been described elsewhere [11][12].
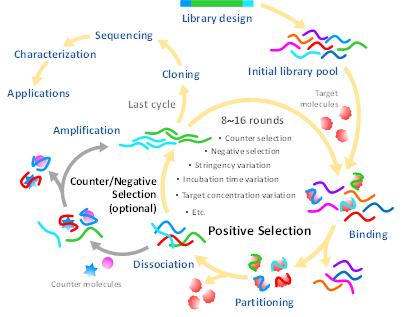
Briefly, SELEX involves the repetitive consecutive steps of binding, partitioning, dissociating and amplifying [13] (Figure 1). In general, each strand of an oligonucleotide library is composed of a random sequence flanked by two known sequences at its 5′ and 3′ termini for specific hybridization with primers during the amplification step. Through positive selection, oligonucleotide sequences are allowed to behave in a native tertiary structure for free recognition of the target, particularly by heating to 90–95 °C and cooling on ice for a period of time (5–10 min) and subsequently incubated with the target molecules. The target-bound oligonucleotides are partitioned from the unbound species and then amplified by polymerase chain reaction (PCR) to increase the copy numbers for execution of the subsequent rounds of the selection process. The successive processes of SELEX are usually carried out over about 8–15 cycles. After the final round of selection, the target-bound oligonucleotides are subjected to sequencing and some essential properties are characterized, such as affinity and selectivity for the target, molecular structure and potential uses. Over the past three decades, the SELEX process has been documented in more than 30 variants and systematically reviewed in many studies [14]; the variants are mainly intended to achieve some of the following: enhanced specificity and affinity, reduced processing time and complexity, expanded target prevalence and improved high-throughput screening.
Figure 1. Typical steps of the systematic evolution of ligands by exponential enrichment (SELEX) procedure.
2. Improvement of Aptamer Specificity and Affinity
Aptamers have limited chemical diversity since they are constructed from a combination of only four nucleobases to produce certain conformations and binding motifs for specific recognition of target molecules. In contrast to the 20 amino acid compositions with their post-translational modifications, antibodies can generate relatively more variations of binding pockets, increasing the possibility to selectively and strongly recognize target molecules than the equivalent oligonucleotide aptamers. Hence, to increase the possibility of acquiring high-affinity and selective aptamers against targets, many strategies have been employed, including but not limited to negative selection, counter selection, stringency amendment, aptamer length optimization and post-selection modification.
3. Essential Characterization of Pesticide Aptamers
Pesticides are much smaller in size than aptamers and reveal very little about their interaction events, making it difficult not only to isolate them but also to characterize their essential properties. Characterization is required to point out the general properties (e.g., secondary and tertiary conformations) and ligand binding performance (e.g., affinity and selectivity) of aptamers, which are important in choosing the right aptamer for the right applications.
Computational analysis is a useful method to illuminate the conformation of the candidate aptamer. The Mfold web server is typically employed to elucidate the secondary structure and relevant Gibbs free energy value (∆G°) of aptamers [15][16][17]. Abraham et al. determined a candidate atrazine-specific aptamer (R12.4) folding by using the Mfold service and found that its structure belonged mostly to a random region and partially to constant regions at the 5′ and 3′ termini. Guided by the predicted structure, truncation of the R12.4 aptamer was initiated by trimming the oligo sequences in both constant and random regions located outside the major hairpin structure in order to reduce cost and increase the efficiency of aptamer synthesis [18].
Furthermore, the secondary structure of aptamers can be experimentally elucidated by circular dichroism (CD) spectroscopy. Conformational variants of aptamers can be distinguished by absorption characteristics of left- and right-polarized light, displayed as unique negative and positive bands of the CD spectrum. For example, a B-form DNA structure shows a negative and a positive band at around 245 and 270 nm, respectively [18]. G-quadruplex structure usually shows a maximum peak at ~260 and a minimum peak at ~240 nm [19]. In addition to its use in exploring the secondary structure, CD is also a valuable tool for revealing the structural changes of aptamers upon binding with ligands [20]. However, the CD spectrum represents only global features of molecules; it cannot provide deep speculation into the binding pocket, which needs to be interpreted with other means, such as computational structure prediction, nuclear magnetic resonance (NMR) or X-ray diffraction, to gain more detailed structural information of the aptamer-ligand complex [21][22].
The aptamer-target affinity is generally determined in terms of the binding constant (KA) or its reciprocal dissociation constant (KD), which can be estimated from the apparent isotherm of aptamer and ligand titration derived by an analytical method such as surface plasmon resonance (SPR), isothermal titration calorimetry (ITC), fluorescence saturation binding assay or capillary electrophoresis (CE). The fluorescence binding assay is simple and requires less sophisticated instruments but needs to label aptamers with fluorescent probes. Commonly, the target molecule is immobilized and incubated with various concentrations of fluorescent-labeled aptamer. After washing out the unbound sequences, the target-bound aptamer is eluted and fluorescence intensity is measured to determine the KD parameter based on an appropriate fitting model of the binding isotherm, such as nonlinear regression analysis [16][23]. In order to circumvent the target immobilization step and avoid aptamer alteration by the labeled fluorophore, molecular beacons can be used as fluorescent probes to generate the binding curve of the aptamer–target complex [24]. This technique allows aptamers to freely interact with targets in their native conformation and lets beacon probes hybridize with free aptamers, which changes the fluorescence emission relative to the complex fraction.
Recently, SPR and ITC have been widely adopted as label-free methods for analyzing biomolecular interactions. SPR functions on the principle that the binding of aptamers with immobilized targets or vice versa, on an SPR chip causes refractive index changes, traced in real time as a sensorgram. SPR provides crucial binding parameters, especially KA, KD, kinetic rate constants (kon and koff) and stoichiometry by estimating from the aptamer–ligand binding isotherm [25][26][27]. ITC directly measures temperature change as a result of the aptamer and target interaction. It elucidates the essential information about thermodynamic parameters of the molecular interaction, such as Gibbs free energy change (∆G°), entropy change (∆S°) and enthalpy change (∆H°), which helps explain the interaction mechanism, major binding forces and spontaneity of the aptamer–ligand complex. Both SPR and ITC provide kinetic information about the interaction mechanism with the requirement of small sample volume, making them applicable for characterization of the aptamer–pesticide duplex [18][25][17][28].
This entry is adapted from the peer-reviewed paper 10.3390/s20236809
References
- Wu, Y.X.; Kwon, Y.J. Aptamers: The “evolution” of SELEX. Methods 2016, 106, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Reverdatto, S.; Burz, D.S.; Shekhtman, A. Peptide aptamers: Development and applications. Curr. Top. Med. Chem. 2015, 15, 1082–1101. [Google Scholar] [CrossRef]
- Beisel, C.L.; Smolke, C.D. Design principles for riboswitch function. PLoS Comput. Biol. 2009, 5, e1000363. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Bock, L.C.; Griffin, L.C.; Latham, J.A.; Vermaas, E.H.; Toole, J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 1992, 355, 564–566. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Cheng, H.; Wang, J.; Xu, L.; Chen, H.; Pei, R. Selection and characterization of DNA aptamers for the development of light-up biosensor to detect Cd(II). Talanta 2016, 154, 498–503. [Google Scholar] [CrossRef]
- Ma, X.; Wang, W.; Chen, X.; Xia, Y.; Wu, S.; Duan, N.; Wang, Z. Selection, identification, and application of Aflatoxin B1 aptamer. Eur. Food Res. Technol. 2014, 238, 919–925. [Google Scholar] [CrossRef]
- Yu, Q.; Liu, M.; Su, H.; Xiao, H.; Wu, S.; Qin, X.; Li, S.; Mi, H.; Lu, Z.; Shi, D.; et al. Selection and characterization of ssDNA aptamers specifically recognizing pathogenic Vibrio alginolyticus. J. Fish Dis. 2019, 42, 851–858. [Google Scholar] [CrossRef]
- Affinito, A.; Quintavalle, C.; Esposito, C.L.; Roscigno, G.; Vilardo, C.; Nuzzo, S.; Vitiani, L.R.; De Luca, G.; Pallini, R.; Kichkailo, A.S.; et al. The discovery of RNA aptamers that selectively bind glioblastoma stem cells. Mol. Ther. Nucleic Acids 2019, 18, 99–109. [Google Scholar] [CrossRef]
- Zhang, Y.; Lai, B.S.; Juhas, M. Recent advances in aptamer discovery and applications. Molecules 2019, 24, 941. [Google Scholar] [CrossRef]
- Stoltenburg, R.; Reinemann, C.; Strehlitz, B. SELEX—A (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 2007, 24, 381–403. [Google Scholar] [CrossRef]
- Wu, Y.; Belmonte, I.; Sykes, K.S.; Xiao, Y.; White, R.J. Perspective on the future role of aptamers in analytical chemistry. Anal. Chem. 2019, 91, 15335–15344. [Google Scholar] [CrossRef]
- Yan, J.; Xiong, H.; Cai, S.; Wen, N.; He, Q.; Liu, Y.; Peng, D.; Liu, Z. Advances in aptamer screening technologies. Talanta 2019, 200, 124–144. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Liu, Y.; Fan, M.; Liu, X. Isolation and identification of the DNA aptamer target to acetamiprid. J. Agric. Food Chem. 2011, 59, 1582–1586. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.M.; Crihfield, C.L.; Gattu, S.; Holland, L.A.; Sooter, L.J. In vitro selection of a single-stranded DNA molecular recognition element against atrazine. Int. J. Mol. Sci. 2014, 15, 14332–14347. [Google Scholar] [CrossRef]
- Kwon, Y.S.; Nguyen, V.-T.; Park, J.G.; Gu, M.B. Detection of iprobenfos and edifenphos using a new multi-aptasensor. Anal. Chim. Acta 2015, 868, 60–66. [Google Scholar] [CrossRef]
- Abraham, K.M.; Roueinfar, M.; Ponce, A.T.; Lussier, M.E.; Benson, D.B.; Hong, K.L. In vitro selection and characterization of a single-stranded DNA aptamer against the herbicide atrazine. ACS Omega 2018, 3, 13576–13583. [Google Scholar] [CrossRef]
- Cai, S.; Yan, J.; Xiong, H.; Liu, Y.; Peng, D.; Liu, Z. Investigations on the interface of nucleic acid aptamers and binding targets. Analyst 2018, 143, 5317–5338. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Zhao, G.; Liu, M.; Fan, L.; Cao, T. Aptamer-based colorimetric sensing of acetamiprid in soil samples: Sensitivity, selectivity and mechanism. J. Hazard. Mater. 2013, 260, 754–761. [Google Scholar] [CrossRef] [PubMed]
- Weisshoff, H.; Wenzel, K.; Schulze-Rothe, S.; Nikolenko, H.; Davideit, H.; Becker, N.-P.; Göttel, P.; Srivatsa, G.S.; Dathe, M.; Müller, J.; et al. Characterization of aptamer BC 007 substance and product using circular dichroism and nuclear magnetic resonance spectroscopy. J. Pharm. Sci. 2018, 107, 2033–2041. [Google Scholar] [CrossRef] [PubMed]
- Cheung, Y.-W.; Kwok, J.; Law, A.W.L.; Watt, R.M.; Kotaka, M.; Tanner, J.A. Structural basis for discriminatory recognition of Plasmodium lactate dehydrogenase by a DNA aptamer. Proc. Natl. Acad. Sci.USA 2013, 110, 15967–15972. [Google Scholar] [CrossRef] [PubMed]
- Hong, K.L.; Sooter, L.J. In vitro selection of a single-stranded DNA molecular recognition element against the pesticide fipronil and sensitive detection in river water. Int. J. Mol. Sci. 2017, 19, 85. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, X.; Zhang, Q.; Zhang, C.; Liu, Y.; Tu, K.; Tu, J. Selection of DNA aptamers that bind to four organophosphorus pesticides. Biotechnol. Lett. 2012, 34, 869–874. [Google Scholar] [CrossRef]
- Cao, F.; Lu, X.; Hu, X.; Zhang, Y.; Zeng, L.; Chen, L.; Sun, M. In vitro selection of DNA aptamers binding pesticide fluoroacetamide. Biosci. Biotechnol. Biochem. 2016, 80, 823–832. [Google Scholar] [CrossRef]
- Oshannessy, D.J.; Brighamburke, M.; Soneson, K.K.; Hensley, P.; Brooks, I. Determination of rate and equilibrium binding constants for macromolecular interactions using surface plasmon resonance: Use of nonlinear least squares analysis methods. Anal. Biochem. 1993, 212, 457–468. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.L.; McKeague, M.; Smolke, C.D. Facile characterization of aptamer kinetic and equilibrium binding properties using surface plasmon resonance. Methods Enzymol 2014, 549, 451–466. [Google Scholar] [CrossRef]
- Nguyen, V.-T.; Kwon, Y.S.; Kim, J.H.; Gu, M.B. Multiple GO-SELEX for efficient screening of flexible aptamers. ChemComm 2014, 50, 10513–10516. [Google Scholar] [CrossRef] [PubMed]