Tumorigenesis is a complex and multistep process in which sequential mutations in oncogenes and tumor-suppressor genes result in enhanced proliferation and apoptosis escape. Over the past decades, several studies have provided evidence that tumors are more than merely a mass of malignant cancer cells, with the tumor microenvironment (TME) also contributing to cancer progression. For this reason, the focus of cancer research in recent years has shifted from the malignant cancer cell itself to the TME and its interactions. Since the TME actively participates in tumor progression, therapeutic strategies targeting it have created great interest. In this context, much attention has been paid to the potential application of small interfering RNA (siRNA), a class of non-coding RNA that has the ability to downregulate the expression of target genes in a sequence-specific way. This is paving the way for a novel therapeutic approach for the treatment of several diseases, including cancer.
- siRNA,RNA,Tumor
1. Introduction
RNAi is a post-transcriptional gene silencing mechanism participating in the natural process of resistance to the invasion of pathogenic, exogenous, double-stranded RNA; it is executed by three types of small non-coding RNAs (microRNA, siRNA, and short hairpin RNA (shRNA)) [1][2]. siRNA and miRNA can knock down the expression of target genes in a sequence-specific way by inducing mRNA degradation (for siRNA and miRNA) or blocking mRNA translation (for miRNA), a kind of post-transcriptional regulation widely investigated in cancer[3][4]. siRNA consists in a double strand of RNA containing a homologous sequence to a specific gene, with the ability to silence it. The process starts in the cytoplasm, where the enzyme Dicer cleaves a double-stranded RNA into a shorter double strand 21–25 nt (siRNA) in length. The guide strand is loaded into the RNA-induced silencing (RISC) complex, and when it recognizes, via a perfect match, a target mRNA, the RISC complex leads to the cleavage of the mRNA. In this manner, siRNA can regulate gene expression, triggering efficient and specific gene silencing. By mimicking this natural process, it is possible to control the expression of specific disease genes, leading to precise and personalized treatments. Several in vitro studies have shown the efficiency of this mechanism in numerous pathologies, including cancer[5].
From a therapeutic point of view, siRNAs have several advantages with respect to small therapeutics drugs and monoclonal antibodies. Indeed, they act through perfect base pairing with mRNA, whereas small molecules and monoclonal antibodies need to recognize the complex tridimensional conformation of a target protein. Furthermore, any gene of interest can be easily targeted by an siRNA, since only a complementary nucleotide sequence on the targeted mRNA is needed to be designed, whereas conventional compounds can target only a subset of proteins/pathways, defined as “druggable” targets.
Although siRNAs are promising, several barriers limit clinical application. Unmodified siRNAs have some disadvantages, such as inadequate stability in the circulation due to the presence of RNAses that may quickly degrade them, a poor pharmacokinetic properties, and the possible induction of off-target effects if an imperfect match is tolerated by the RISC, leading to undesirable silencing of genes[6]. Moreover, siRNA can trigger the activation of Toll-like receptor 3 (TLR3), negatively influencing the immune system. The occurrence of side effects of siRNA has thus raised many doubts on their possible clinical use. Indeed, siRNA-based therapeutics has suffered many ups and downs since the discovery of siRNA in 1998. To maximize treatment efficacy and reduce the side effects of siRNA, researchers have focused much effort in recent years on the identification of chemical modifications that would aid the development of delivery systems[7].
Interestingly, Patisiran has been recently approved by FDA as the first siRNA therapeutic for the treatment of hereditary amyloidogenic transthyretin amyloidosis. Givosiran has also been recently approved by the FDA for the treatment of acute hepatic porphyria[8][9][10][11]. Thus, siRNA technology may also be promising for the treatment of several other diseases, such as cancer. There are no RNAi-based drugs approved for anticancer therapy, but several therapeutics are currently undergoing clinical trials. Clinical trial involving Ephrin type-A receptor 2 (EphA2) and M2 subunit of ribonucleotide reductase (R2) siRNA-mediated therapy are currently ongoing for Hepatocellular carcinoma (HCC)patients and solid tumors, respectively [12][13][14].
2. siRNA and Breast Cancer
Breast cancer remains the leading cause of cancer-related death and the most frequently diagnosed non-cutaneous malignancy in women worldwide. Most breast cancers are the result of genetic mutations originating in the epithelial cells forming the mammary gland. However, tumor development is also accompanied by changes in the surrounding environment. According to the tumor type, current treatment options include chemotherapeutics, hormonal, and antibody approaches, which render an overall survival of 5 years. Nevertheless, patients can develop tumor relapse and metastasis, and for this reason research in these fields has increased considerably. One extensively explored alternative approach is based on siRNAs: they can be highly selective and target different molecular pathways in breast cancer to influence the crosstalk with the tumor microenvironment. By simply customizing the sequence, a siRNA can be used to target a gene encoding a specific protein isotype and silence the gene expression of different TME cells that compose the tumor mass.
Furthermore, a cocktail of siRNAs can downregulate several pathways simultaneously in an easier manner than small molecule inhibitors or antibodies, which can only target “druggable” proteins.
Unfortunately, siRNA can have off-target effects, and for this reason the avoidance of nonspecific toxicity is a major goal in the development of siRNA-based therapy. The toxicity of siRNA is determined by the presence of complementary sequences in the transcriptome. Therefore, extensive analyses and careful preclinical tests must be performed for a given siRNA prior to use for therapeutic purposes.
The effectiveness of siRNA depends on the ability to reach the target. In a complex in vivo system, the obstacles are exacerbated by the complexity of the animal body at different levels, such as the negative charge of the cell membrane, instability in blood, poor pharmacokinetics, rapid clearance from the blood through liver accumulation and renal filtration, and Toll-like immune responses. For this reason, great effort is directed at improving delivery of siRNA.
The development of siRNA carriers is particularly important when targeting the tumor microenvironment. Breast cancer is known to contain abundant TME cells that can exacerbate pathways at the basis of the tumor progression, escape, and anti-tumor immune response. SiRNA-based therapeutics targeting the pro-tumoral TME can help to hinder the dangerous crosstalk between cancer and TME cells (Figure 1).
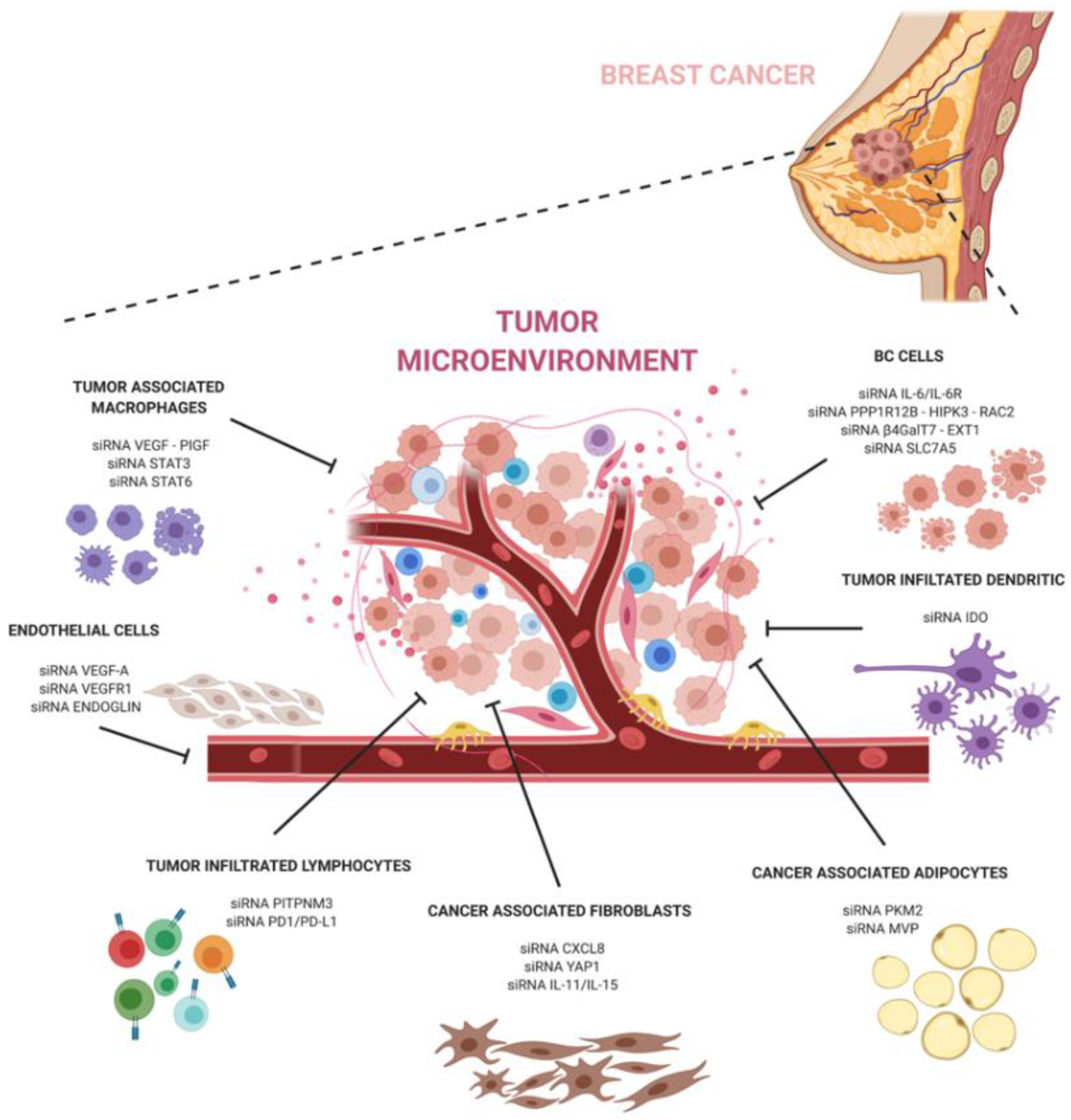
In conclusion, siRNA technology holds great promise in cancer therapy and, together with a better understanding of cancer and its microenvironment interactions, can have a great potential to revolutionize breast cancer treatment and significantly improve the outcomes of breast cancer patients.
This entry is adapted from the peer-reviewed paper 10.3390/cancers12123744
References
- Ryan L. Setten; John J. Rossi; Si-Ping Han; The current state and future directions of RNAi-based therapeutics. Nature Reviews Drug Discovery 2019, 18, 421-446, 10.1038/s41573-019-0017-4.
- C. Napoli; C. Lemieux; R. Jorgensen; Introduction of a Chimeric Chalcone Synthase Gene into Petunia Results in Reversible Co-Suppression of Homologous Genes in trans.. The Plant Cell 1990, 2, 279-289, 10.1105/tpc.2.4.279.
- Giuseppina Roscigno; Assunta Cirella; Alessandra Affinito; Cristina Quintavalle; Iolanda Scognamiglio; Francesco Palma; Francesco Ingenito; Silvia Nuzzo; Francesca De Micco; Antonio Cuccuru; et al. miR-216a Acts as a Negative Regulator of Breast Cancer by Modulating Stemness Properties and Tumor Microenvironment. International Journal of Molecular Sciences 2020, 21, 2313, 10.3390/ijms21072313.
- Giuseppina Roscigno; Ilaria Puoti; Immacolata Giordano; Elvira Donnarumma; Valentina Russo; Alessandra Affinito; Assunta Adamo; Cristina Quintavalle; Matilde Todaro; Maria Dm Vivanco; et al. MiR-24 induces chemotherapy resistance and hypoxic advantage in breast cancer. Oncotarget 2017, 8, 19507-19521, 10.18632/oncotarget.14470.
- Xavier M. Anguela; Katherine A. High; Entering the Modern Era of Gene Therapy. Annual Review of Medicine 2019, 70, 273-288, 10.1146/annurev-med-012017-043332.
- Won Gil Cho; Romulo J. C. Albuquerque; Mark E. Kleinman; Valeria Tarallo; Adelaide Greco; Miho Nozaki; Martha G. Green; Judit Z. Baffi; Balamurali K. Ambati; Massimo De Falco; et al. Small interfering RNA-induced TLR3 activation inhibits blood and lymphatic vessel growth. Proceedings of the National Academy of Sciences 2009, 106, 7137-7142, 10.1073/pnas.0812317106.
- Sayda M. Elbashir; Jens Harborth; Winfried Lendeckel; Abdullah Yalcin; Klaus Weber; Thomas Tuschl; Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001, 411, 494-498, 10.1038/35078107.
- Eliane Sardh; Pauline Harper; Manisha Balwani; Penelope Stein; David Rees; D. Montgomery Bissell; Robert Desnick; Charles Parker; John Phillips; Herbert L. Bonkovsky; et al. Phase 1 Trial of an RNA Interference Therapy for Acute Intermittent Porphyria. New England Journal of Medicine 2019, 380, 549-558, 10.1056/nejmoa1807838.
- Manisha Balwani; Eliane Sardh; Paolo Ventura; Paula Aguilera Peiró; David C. Rees; Ulrich Stölzel; D. Montgomery Bissell; Herbert L. Bonkovsky; Jerzy Windyga; Karl E. Anderson; et al. Phase 3 Trial of RNAi Therapeutic Givosiran for Acute Intermittent Porphyria. New England Journal of Medicine 2020, 382, 2289-2301, 10.1056/nejmoa1913147.
- Pedro Renato De Paula Brandão; Simoneide S. Titze-De-Almeida; Ricardo Titze-De-Almeida; Leading RNA Interference Therapeutics Part 2: Silencing Delta-Aminolevulinic Acid Synthase 1, with a Focus on Givosiran. Molecular Diagnosis & Therapy 2019, 24, 61-68, 10.1007/s40291-019-00438-6.
- Sagar Agarwal; Amy R. Simon; Varun Goel; Bahru A. Habtemariam; Valerie A. Clausen; Jae B. Kim; Gabriel J. Robbie; Pharmacokinetics and Pharmacodynamics of the Small Interfering Ribonucleic Acid, Givosiran, in Patients With Acute Hepatic Porphyria. Clinical Pharmacology & Therapeutics 2020, 108, 63-72, 10.1002/cpt.1802.
- Jeremy D. Heidel; Joanna Yi-Ching Liu; Yun Yen; Bingsen Zhou; Bret S.E. Heale; John J. Rossi; Derek W. Bartlett; Mark E Davis; Potent siRNA Inhibitors of Ribonucleotide Reductase Subunit RRM2 Reduce Cell Proliferation In vitro and In vivo. Clinical Cancer Research 2007, 13, 2207-2215, 10.1158/1078-0432.ccr-06-2218.
- Jeremy D. Heidel; Zhongping Yu; Joanna Yi-Ching Liu; Shyam M. Rele; Yongchao Liang; Ryan K. Zeidan; Douglas J. Kornbrust; Mark E Davis; Administration in non-human primates of escalating intravenous doses of targeted nanoparticles containing ribonucleotide reductase subunit M2 siRNA. Proceedings of the National Academy of Sciences 2007, 104, 5715-5721, 10.1073/pnas.0701458104.
- Michael J. Wagner; Rahul Mitra; Mark J. McArthur; Wallace Baze; Kirstin Barnhart; Sherry Y. Wu; Cristian Rodriguez-Aguayo; Xinna Zhang; Robert L. Coleman; Gabriel Lopez-Berestein; et al. Preclinical Mammalian Safety Studies of EPHARNA (DOPC Nanoliposomal EphA2-Targeted siRNA). Molecular Cancer Therapeutics 2017, 16, 1114-1123, 10.1158/1535-7163.mct-16-0541.
