Genetic code symmetries in form of the Supersymmetry Genetic Code (SSYGC) table support the wobble hypothesis with non-Watson–Crick pairing interactions between the translation process from mRNA to tRNA. Namely, in 1966 F.H.C. Crick proposed the wobble hypothesis to explain this partial degeneracy. Without symmetries of genetic code wobbling interaction can have misreading error in proteinogenesis.
- codons
- mRNA
- tRNA
- aaRSs
- wobble hypothesis
- genetic code symmetry
- the supersymmetry genetic code table
1. Introduction
Genetic code symmetries in form of the Supersymmetry Genetic Code (SSYGC) [1] table support the wobble hypothesis with non-Watson–Crick pairing interactions between the translation process from mRNA to tRNA (Figure 1). Namely, in 1966 F.H.C. Crick proposed the wobble hypothesis to explain this partial degeneracy [2]. Without symmetries of genetic code wobbling interaction can have misreading error in proteinogenesis.
The tRNA molecule is the bridging molecule and the link between mRNA and aaRSs which play a crucial role in translation as an essential and universally distributed family of enzymes, pairing tRNA with their cognate amino acids to decode mRNA according to the genetic code [3][4]. Cognate amino acid corresponds to the anticodon triplet of the tRNA according to the genetic code. The aaRSs as the group of enzymes are capable of implementing the genetic code [5]. It means, the correct translation ensures that a cognate amino acid is under the control of aaRSs and under genetic code symmetries control.
Only tRNAs with correct anticodon are bound tightly by the ribosome. The whole tRNA molecule has 75 bases. The anticodon stem and loop domain contain three bases on position 34, 35 and 36. The direction of codons in mRNA is 5’3’ due to ester binding between phosphate 5’ and sugar 3’. The third base of the codon has a 3’ end position. The tRNA anticodon has an antiparallel direction with a 5’ end position of the first anticodon base on the anticodon stem and loop domain of the tRNA molecule (Figure 2). Namely, it is very important to emphasise that the first and second nucleotides in the codon opposite the second and third nucleotides in the anticodon are always strictly positioned according to canonical Watson–Crick complementary pairing with A↔U and C↔G bases. Consequently, the second base has the same central position in the codon and anticodon. The role of the second base in the codons of the SSyGC table reveals which codons belong to amino acids with the whole non-split box of four codons, and which belong to amino acids with two codons each from split boxes [6]. The second base in mRNA codons can have an identical role. This information transfers the tRNA anticodon to aaRSs. When a tRNA reaches an aaRS synthetases bind its amino acid to own acceptor stem t-shaped RNA.
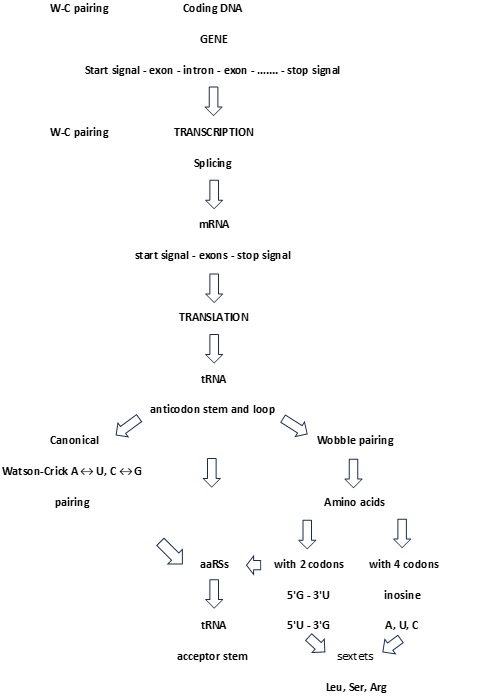
Figure 1. Algorithm of proteinogenesis. All stages from coding DNA over mRNA to tRNA are based on Watson-Crick pairing. The critical point is the process of translation because the number of mRNA codons is larger than the number of tRNA anticodons. As a result, canonical base pairing must be supplemented with wobble non-canonical hydrogen bonding 5’G→3’U or 5’U→3’G for an amino acid with two codons, and inosine A, inosine U, inosine C for amino acids with four codons. In this case, the SSyGC table plays an important role as it discovers the core purine-pyrimidine symmetry net in which each codon has a strictly defined position with respect to other codons. At the same time, each codon with its purine-pyrimidine structure of first and second bases reveals whether it is from an amino acid with two or four codons.
2. The Relationship between the SSyGC Table and Wobble Hypothesis
The tRNA molecule can recognise a different first base at the 5’ position of the anticodon as wobble pairing, owing to a non-canonical Watson–Crick base pair with the mRNA third base at the 3’ position in the codon–anticodon interaction. Namely, there are some differences in the total numbers of codons and tRNAs: for different species, there are on average 40 tRNAs as opposed to the necessary 61 codons and stop signals UAG, UAA and UGA 13]. Wobble pairing partially explains and corrects this disagreement. and minimises damage that can be caused by a misreading of the code [7].
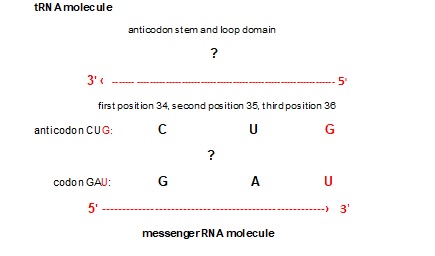
Figure 2. Connection between mRNA and tRNA in translation process of proteinogenesis (example with codon GAU). The anticodon stem and loop domain contain three bases on position 34, 35 [8]and 36 of total 75 bases of tRNA clover shaped molecule. The direction of codons in mRNA is 5’3’ due to ester binding between phosphate 5’ and sugar 3’. The third base of the codon has a 3’ end position. The tRNA anticodon has an antiparallel direction with a 5’ end position of the first anticodon base on the anticodon stem and loop domain of the tRNA molecule. Beside the codon-anticodon canonical Watson-Crick pairing G↔C and U↔A (black), wobble tolerates pairing between e.g. 5’ G anticodon ↔ 3’ U codon (red).
Wobble bases
tRNA C A G U I
mRNA G U C A C
U G A
U
Example 1. tRNA anticodon UUG (5’) wobble base G
mRNA codon AAC canonical base pair
mRNA codon AAU (3’) wobble pairing G-U
AAC and AAU are both codons of Asparagine (Asn) from the split box.
Beside the codon-anticodon canonical Watson-Crick pairing G↔C and U↔A, wobble tolerates pairing between 5’ G anticodon ↔ 3’ U codon in amino acids with two codons and with a third base pyrimidine C and U from split box. From the tRNA anticodon UUG, canonical pairing is codon AAC in mRNA. The wobble pairing is AAU. We show that two first weak bases (AA) in the codon reveal the split box which contains two codons for two different amino acids (Figure 2). In this example, the canonical codon is AAC with pyrimidine C as the third base. However, the wobble allows pairing with AAU, which also has pyrimidine U as a third base, and both codons belong to the same amino acid Asparagine. Due to the symmetries, wobble pairing with the anticodon UUG recognises a second codon, except the regular canonical, from its own amino acid Asparagine. In this way, the message from tRNA is correct and aaRSs for Asparagine can also recognise codons for its amino acid and charged tRNA appropriate amino acid into a growing polypeptide chain. The combination tRNA G → mRNA U, through wobble pairing with only one anticodon, solves the problem regarding an amino acid with two codons both of which have pyrimidines (U, C) as a third base.
Example 2. tRNA anticodon UCU (5) wobble base U
mRNA codon AGA canonical base pair
mRNA codon AGG (3’) wobble pairing U-G
AGA and AGG are codons (2/6) of the sextet Arginine (Arg) from the split box.
The tRNA anticodon UCU has a canonical pairing with codon AGA on mRNA with mixed first two bases (AG), but the second base is purine and therefore the box is split (Figure 2). This amino acid is Arginine with two codons (2/6) from the split box. In other words, the canonical codon is AGA with purine (A) as the third base. The wobble allows pairing with AGG, whose codon also has purine (G) as the third base. AGG and AGA are both codons (2/6 from the split box) for the amino acid Arginine, and the message will be correctly recognised by aaRSs. In this example, the combination anticodon tRNA U → codon mRNA G through wobble pairing solves the problem regarding amino acids with two codons from the split box which have purine (A, G) as the third base. The message from tRNA is correct and aaRSs for the two codons from the split box of Arginine can recognise codon for its amino acid and the charged tRNA appropriate amino acid into a growing polypeptide chain. The remaining four codons (4/6) from the non-split box has wobble pairing with inosine (I) (see example 3).
Example 3. tRNA anticodon AAI (5’) wobble base I
mRNA codons UUA (3’) wobble pairing with H-A
UUC (3’) wobble pairing with H-C
UUU (3’) wobble pairing with H-U
UUA, UUC and UUU are codons of sextet Leucine (Leu) from the non-split box.
Only tRNA can have inosine as the first base (5’) on the anticodon. The nucleobase of inosine is hypoxanthine. There are three types: hypoxanthine adenine (H-A), hypoxanthine cytosine (H-C) and hypoxanthine uracil (H-U). Inosine on the position of first base in anticodon can be matched with three bases A, C and U as the third base (3’) in the codons of mRNA. The wobble with inosine is suitable for amino acids which contain four codons of the non-split box, and the wobble pairing is possible with each of the bases A, C and U. In non-split boxes, the third base G of codon from mRNA always has canonical pairing with the anticodon base C from tRNA.
In our example, the tRNA anticodon AAI has wobble pairing with codons which has the first two weak bases UU, but the second base is pyrimidine U, and therefore this amino acid is from the non-split box and has four codons (Figure 2). Wobble with inosine enables pairing with UUA, UUC and UUU codons in mRNA. The message for all three codons will be appropriate because it belongs to the same amino acid which will be recognised by aaRSs for Leucine. The fourth codon UUG has a canonical Warson-Crick pairing with the AAC anticodon on tRNA. In the genetic code, there are eight amino acids which possess the whole box with four codons, including the sextets Serine, Arginine and Leucine.
These three examples of wobble pairing cover all natural amino acids from the genetic code. It is important to observe that the symmetry modification of the anticodon stem and loop domain on tRNA with wobble pairing include the positions 34, 35 and 36 with the first base of the anticodon in position 34. Because of less tRNA opposite mRNA, wobble pairing ensures that not one amino acid remains out of the translation process.
A question is why does only inosine satisfy wobble pairing with an amino acid from the non-split box? See Example 1 for the two codons AAC and AAU from the amino acid Asparagine from the split box. The wobble pairing with the anticodon inosine UUI in this example could also recognise the AAA codon from the second amino acid Lysine from each box of mRNA as a misreading error.
Example 2 for the two codons AGA and AGG from the split box of amino acid Arginine show that wobble pairing is possible with the inosine UCI anticodon for the AGA codon. In this example, inosine could also recognise AGC and AGU codons from the split box of the second amino acid Serine as a misreading error. Wobble pairing with inosine is correct and without misreading, with only 8 amino acids which have four codons from the non-split box.
Concluding, the misreading errors in the translation process of proteinogenesis are prevented due to the genetic code symmetries, which enable the recognition of each codon whether it belongs to amino acid coded by two codons from the split box, or by all four codons from the non-split box what is important for the correct wobble pairing [8]. The role of physicochemical purine–pyrimidine symmetries of genetic code is to decrease disorder according to error minimisation and preserve the integrity of biological processes during proteinogenesis.
References
- Rosandić, M.; Paar, V.; Standard Genetic code vs. Supersymmetry Genetic Code–Alphabetical table vs. physicochemical table. BioSystems 2022, 14, 110748 , .
- Crick, F.H.C.; Standard Genetic code vs. Supersymmetry Genetic Code–Alphabetical table vs. physicochemical table. J. Mo. Biol. 1966, 19, 548-555 , .
- Ibba, M.; Soll, D.; Aminoacyl-tRNA synthetases. Annu Rev. Biochem. 2000, 69, 617-650 , .
- Gomez, M. A. R.; Ibba, M.; Aminoacyl tRNA synthetases. RNA 2020, 26(8), 910-936 , .
- Woese, C. R.; Olsen, G. J.; Ibba, M.; Söll., D.; Aminoacyl synthetases, the genetic code, and the evolutionary process. . Microbiol. Mol. Biol. Rev. 2000, 64, 202-236 , .
- Rosandić, M.; Paar, V.; Maximal Genetic Code Symmetry Is a Physicochemical Purine-Pyrimidine Symmetry Language for Transcription and Translation in the Flow of Genetic Information from DNA to Protein. Int. J. Mol. Sci. 2024, 25, 9543, .
- Agris, P. F.; Eruysal, E. R.; Narendran, A.; Väre, V. Y. P.; Vangavati, S.; Ranganathan, S., V.; Celebrating wobble decoding: Half a century and still much is new. RNA Biol. 2018, 14(4-5), 537-553, .
- Rosandic, M. Translation in Proteinogenesis Based on Genetic Code symmetry. Scholarly Community Encyclopedia 2025.
