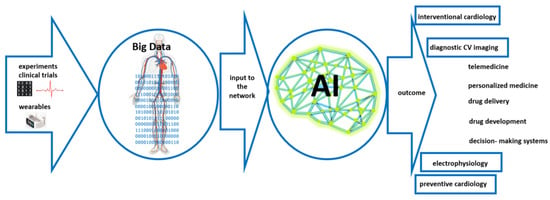
Artificial Intelligence (AI)-based algorithms, in particular, Deep Neural Networks (DNNs), have recently revolutionized image creation. Precise segmentation of lesions may contribute to an efficient diagnostics process and a more effective selection of targeted therapy. For example, an AI-based algorithm for the segmentation of pigmented skin lesions has been developed, which enables diagnosis in the earlier stages of the disease, without invasive medical procedures. With flexibility and scalability, AI can be also considered an efficient tool for cancer diagnosis, particularly in the early stages of the disease.
- Artificial Intelligence
- Machine Learning
- Virtual Reality
- Extended Reality
- Mixed Reality
- cardiology
- personalized medicine
- Metaverse
- Augmented Reality
1. Introduction
| AI/ML Model | Application Fields (In General) | Application Fields (In Cardiology) | References |
|---|---|---|---|
| ANNs | classification, pattern recognition, image recognition, natural language processing (NLP), speech recognition, recommendation systems, prediction, cybersecurity, object manipulation, path planning, sensor fusion | prediction of atrial fibrillation, acute myocardial infarctions, and dilated cardiomyopathy detection of the structural abnormalities in heart tissues | [8] [9] |
| RNNs | ordinal or temporal problems (language translation, speech recognition, NLP image captioning), time series prediction, music generation, video analysis, patient monitoring, disease progression prediction | segmentation of the heart and subtle structural changes cardiac MRI segmentation |
[10] [11] |
| LSTMs | ordinal or temporal problems (language translation, speech recognition, NLP, image captioning), time series prediction, music generation, video analysis, patient monitoring, disease progression prediction | segmentation and classification of 2D echo images segmentation and classification of 3D Doppler images segmentation and classification of video graphics images and detection of the AMI in echocardiography |
[12] [13] |
| CNNs | pattern recognition, segmentation/classification, object detection, semantic segmentation, facial recognition, medical imaging, gesture recognition, video analysis | cardiac image segmentation to diagnose CAD cardiac image segmentation to diagnose Tetralogy of Fallot localization of the coronary artery atherosclerosis detection of cardiovascular abnormalities detection of arrhythmia detection of coronary artery disease prediction of the survival status of heart failure patients prediction of cardiovascular disease LV dysfunction screening prediction of premature ventricular contraction detection |
[14][15] [16] [17] [18] [19][20][21][22][23][24][25][26][27][28][29][30] [31] [32] [33] [34][35] |
| Transformers | NLP, speech processing, computer vision, graph-based tasks, electronic health records, building conversational AI systems and chatbots | coronary artery labeling prediction of incident heart failure arrhythmia classification cardiac abnormality detection segmentation of MRI in case of cardiac infarction classification of aortic stenosis severity LV segmentation heart murmur detection myocardial fibrosis segmentation ECG classification |
[36][37] [38] [39][40][41][42] [43] [44] [45][46] [41][47][48] [49] [41] [50] |
| SNNs | pattern recognition, cognitive robotics, SNN hardware, brain–machine interfaces, neuromorphic computing | ECG classification detection of arrhythmia extraction of ECG features |
[51][52][53] [54][55][56] [57] |
| GANs | image-to-image translation, image synthesis, and generation, data generation for training, data augmentation, creating realistic scenes | CVD diagnosis segmentation of the LA and atrial scars in LGE CMR images segmentation of ventricles based on MRI scans left ventricle segmentation in pediatric MRI scans generation of synthetic cardiac MRI images for congenital heart disease research |
[58] [59] [60] [61] [62] |
| GNNs | graph/node classification, link prediction, graph generation, social/biological network analysis, fraud detection, recommendation systems | classification of polar maps in cardiac perfusion imaging analysis of CT/MRI scans prediction of ventricular arrhythmia segmentation of cardiac fibrosis diagnosis of cardiac condition: LV motion in cardiac MR cine images automated anatomical labeling of coronary arteries prediction of CAD automation of coronary artery analysis using CCTA screening of cardio, thoracic, and pulmonary conditions in chest radiograph |
[63][64] [65] [64] [64] [66] [67] [68] [69] [70] |
| QNNs | optimization of hardware operations, user interfaces | classification of ischemic heart disease | [20] |
| GA | optimization techniques, risk prediction, gene therapies, medicine development | classification of heart disease | [71] |
2. Artificial Intelligence-Based Support in Cardiology
2.1. Application of the You-Only-Look-Once (YOLO) Algorithm
2.2. Genetic Algorithms
2.3. Artificial Neural Networks
2.4. Convolutional Neural Networks
2.5. Recurrent Neural Networks
2.6. Spiking Neural Networks
2.7. Generative Adversarial Networks
2.8. Graph Neural Networks
2.9. Transformers
2.10. Quantum Neural Networks
3. Evaluation Metrics in Medical Image Segmentation
Moreover, an important element in improving the effectiveness of cardiology data segmentation is the collection of as much reliable, good-quality data as possible while keeping class balance in mind. This procedure should take into account input data diversity that helps AI models better generalize unseen cases while their reliability is improved. It is also necessary to provide diverse and representative input data whenever possible, which can help mitigate bias in AI-based algorithms. Another issue related to data is the application of the open data policy following UNESCO guidelines (especially for scientific applications, and research) so that more efficient AI algorithms can be developed in the area of cardiology. Moreover, compliance with ethical and bioethical standards in the collection, storage, and use of medical data is essential for the development of reliable AI systems in cardiology. As a consequence, the establishment of standards for the quality, integrity, and interoperability of cardiology data used in AI applications in cardiology as well as the development of the protocols for the validation and regulation of AI-based algorithms is of high importance. It is also necessary to develop guidelines on how to integrate artificial intelligence technologies into cardiology workflows as well as strategies for managing risks associated with the implementation of AI-based technologies in cardiology. Finally, it should be the responsibility of the cardiology community to ensure the control of results and feedback loops in the implementation of mechanisms for monitoring the performance of AI algorithms in cardiology and the collection of feedback from clinicians and patients.
This entry is adapted from the peer-reviewed paper 10.3390/electronics13050866
References
- Boonstra, M.J.; Oostendorp, T.F.; Roudijk, R.W.; Kloosterman, M.; Asselbergs, F.W.; Loh, P.; Van Dam, P.M. Incorporating Structural Abnormalities in Equivalent Dipole Layer Based ECG Simulations. Front. Physiol. 2022, 13, 2690.
- He, B.; Hu, W.; Zhang, K.; Yuan, S.; Han, X.; Su, C.; Zhao, J.; Wang, G.; Wang, G.; Zhang, L. Image Segmentation Algorithm of Lung Cancer Based on Neural Network Model. Expert Syst. 2021, 39, e12822.
- Evans, L.M.; Sözümert, E.; Keenan, B.E.; Wood, C.E.; du Plessis, A. A Review of Image-Based Simulation Applications in High-Value Manufacturing. Arch. Comput. Methods Eng. 2023, 30, 1495–1552.
- Arafin, P.; Billah, A.M.; Issa, A. Deep Learning-Based Concrete Defects Classification and Detection Using Semantic Segmentation. Struct. Health Monit. 2024, 23, 383–409.
- Ye-Bin, M.; Choi, D.; Kwon, Y.; Kim, J.; Oh, T.H. ENInst: Enhancing Weakly-Supervised Low-Shot Instance Segmentation. Pattern. Recognit. 2024, 145, 109888.
- Hong, F.; Kong, L.; Zhou, H.; Zhu, X.; Li, H.; Liu, Z. Unified 3D and 4D Panoptic Segmentation via Dynamic Shifting Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2024, 1–16.
- Rudnicka, Z.; Szczepanski, J.; Pregowska, A. Artificial Intelligence-Based Algorithms in Medical Image Scan Seg-Mentation and Intelligent Visual-Content Generation-a Concise over-View. Electronics 2024, 13, 746.
- Sammani, A.; Bagheri, A.; Van Der Heijden, P.G.M.; Te Riele, A.S.J.M.; Baas, A.F.; Oosters, C.A.J.; Oberski, D.; Asselbergs, F.W. Automatic Multilabel Detection of ICD10 Codes in Dutch Cardiology Discharge Letters Using Neural Networks. NPJ Digit. Med. 2021, 4, 37.
- Muscogiuri, G.; Van Assen, M.; Tesche, C.; De Cecco, C.N.; Chiesa, M.; Scafuri, S.; Guglielmo, M.; Baggiano, A.; Fusini, L.; Guaricci, A.I.; et al. Review Article Artificial Intelligence in Coronary Computed Tomography Angiography: From Anatomy to Prognosis. BioMed Res. Int. 2020, 2020, 6649410.
- Yasmin, F.; Shah, S.M.I.; Naeem, A.; Shujauddin, S.M.; Jabeen, A.; Kazmi, S.; Siddiqui, S.A.; Kumar, P.; Salman, S.; Hassan, S.A.; et al. Artificial Intelligence in the Diagnosis and Detection of Heart Failure: The Past, Present, and Future. Rev. Cardiovasc. Med. 2021, 22, 1095–1113.
- Samieiyeganeh, M.E.; Rahmat, R.W.; Khalid, F.B.; Kasmiran, K.B. An overview of deep learning techniques in echocardiography image segmentation. J. Theor. Appl. Inf. Technol. 2020, 98, 3561–3572.
- Wahlang, I.; Kumar Maji, A.; Saha, G.; Chakrabarti, P.; Jasinski, M.; Leonowicz, Z.; Jasinska, E.; Dimauro, G.; Bevilacqua, V.; Pecchia, L. Electronics Article. Electronics 2021, 10, 495.
- Muraki, R.; Teramoto Id, A.; Sugimoto, K.; Sugimoto, K.; Yamada, A.; Watanabe, E. Automated Detection Scheme for Acute Myocardial Infarction Using Convolutional Neural Network and Long Short-Term Memory. PLoS ONE 2022, 17, e0264002.
- Roy, S.S.; Hsu, C.H.; Samaran, A.; Goyal, R.; Pande, A.; Balas, V.E. Vessels Segmentation in Angiograms Using Convolutional Neural Network: A Deep Learning Based Approach. CMES Comput. Model. Eng. Sci. 2023, 136, 241–255.
- Liu, J.; Yuan, G.; Yang, C.; Song, H.; Luo, L. An Interpretable CNN for the Segmentation of the Left Ventricle in Cardiac MRI by Real-Time Visualization. CMES Comput. Model. Eng. Sci. 2023, 135, 1571–1587.
- Tandon, A. Artificial Intelligence in Pediatric and Congenital Cardiac Magnetic Resonance Imaging. In Intelligence-Based Cardiology and Cardiac Surgery: Artificial Intelligence and Human Cognition in Cardiovascular Medicine; Academic Press: London, UK, 2024; pp. 201–209.
- Candemir, S.; White, R.D.; Demirer, M.; Gupta, V.; Bigelow, M.T.; Prevedello, L.M.; Erdal, B.S. Automated Coronary Artery Atherosclerosis Detection and Weakly Supervised Localization on Coronary CT Angiography with a Deep 3-Dimensional Convolutional Neural Network. Comput. Med. Imaging Graph. 2020, 83, 101721.
- Singh, N.; Gunjan, V.K.; Shaik, F.; Roy, S. Detection of Cardio Vascular Abnormalities Using Gradient Descent Optimization and CNN. Health Technol. 2024, 14, 155–168.
- Banerjee, D.; Dey, S.; Pal, A. An SNN Based ECG Classifier for Wearable Edge Devices. In Proceedings of the NeurIPS 2022 Workshop on Learning from Time Series for Health, New Orleans, LA, USA, 2 December 2022.
- Ullah, U.; García, A.; Jurado, O.; Diez Gonzalez, I.; Garcia-Zapirain, B. A Fully Connected Quantum Convolutional Neural Network for Classifying Ischemic Cardiopathy. IEEE Access 2022, 10, 134592–134605.
- Çınar, A.; Tuncer, S.A. Classification of Normal Sinus Rhythm, Abnormal Arrhythmia and Congestive Heart Failure ECG Signals Using LSTM and Hybrid CNN-SVM Deep Neural Networks. Comput. Methods Biomech. Biomed. Engin. 2021, 24, 203–214.
- Fradi, M.; Khriji, L.; Machhout, M. Real-Time Arrhythmia Heart Disease Detection System Using CNN Architecture Based Various Optimizers-Networks. Multimed. Tools Appl. 2022, 81, 41711–41732.
- Rahul, J.; Sharma, L.D. Automatic Cardiac Arrhythmia Classification Based on Hybrid 1-D CNN and Bi-LSTM Model. Biocybern. Biomed. Eng. 2022, 42, 312–324.
- Ahmed, A.A.; Ali, W.; Abdullah, T.A.A.; Malebary, S.J.; Yao, Y.; Huang, X.; Ahmed, A.A.; Ali, W.; Abdullah, T.A.A.; Malebary, S.J. Citation: Classifying Cardiac Arrhythmia from ECG Signal Using 1D CNN Deep Learning Model. Mathematics 2023, 11, 562.
- Eltrass, A.S.; Tayel, M.B.; Ammar, A.I. A New Automated CNN Deep Learning Approach for Identification of ECG Congestive Heart Failure and Arrhythmia Using Constant-Q Non-Stationary Gabor Transform. Biomed. Signal Process. Control 2021, 65, 102326.
- Zheng, Z.; Chen, Z.; Hu, F.; Zhu, J.; Tang, Q.; Liang, Y. Electronics Article. Electronics 2020, 9, 121.
- Cheng, J.; Zou, Q.; Zhao, Y. ECG Signal Classification Based on Deep CNN and BiLSTM. BMC Med. Inf. Decis. Mak. 2021, 21, 365.
- Rawal, V.; Prajapati, P.; Darji, A. Hardware Implementation of 1D-CNN Architecture for ECG Arrhythmia Classification. Biomed. Signal Process. Control 2023, 85, 104865.
- Zhang, Y.; Liu, S.; He, Z.; Zhang, Y.; Wang, C. A CNN Model for Cardiac Arrhythmias Classification Based on Individual ECG Signals. Cardiovasc. Eng. Technol. 2022, 13, 548–557.
- Khozeimeh, F.; Sharifrazi, D.; Izadi, N.H.; Hassannataj Joloudari, J.; Shoeibi, A.; Alizadehsani, R.; Tartibi, M.; Hussain, S.; Sani, Z.A.; Khodatars, M.; et al. RF-CNN-F: Random Forest with Convolutional Neural Network Features for Coronary Artery Disease Diagnosis Based on Cardiac Magnetic Resonance. Sci. Rep. 2022, 12, 17.
- Aslan, M.F.; Sabanci, K.; Durdu, A. A CNN-Based Novel Solution for Determining the Survival Status of Heart Failure Patients with Clinical Record Data: Numeric to Image. Biomed. Signal Process. Control 2021, 68, 102716.
- Yoon, T.; Kang, D. Bimodal CNN for Cardiovascular Disease Classification by Co-Training ECG Grayscale Images and Scalograms. Sci. Rep. 2023, 13, 2937.
- Sun, L.; Shang, D.; Wang, Z.; Jiang, J.; Tian, F.; Liang, J.; Shen, Z.; Liu, Y.; Zheng, J.; Wu, H.; et al. MSPAN: A Memristive Spike-Based Computing Engine With Adaptive Neuron for Edge Arrhythmia Detection. Front. Neurosci. 2021, 15, 761127.
- Wang, J.; Zang, J.; Yao, S.; Zhang, Z.; Xue, C. Multiclassification for Heart Sound Signals under Multiple Networks and Multi-View Feature. Measurement 2024, 225, 114022.
- Wang, L.-H.; Ding, L.-J.; Xie, C.-X.; Jiang, S.-Y.; Kuo, I.-C.; Wang, X.-K.; Gao, J.; Huang, P.-C.; Patricia, A.; Abu, A.R. Automated Classification Model With OTSU and CNN Method for Premature Ventricular Contraction Detection. IEEE Access 2021, 9, 156581–156591.
- Jungiewicz, M.; Jastrzębski, P.; Wawryka, P.; Przystalski, K.; Sabatowski, K.; Bartuś, S. Vision Transformer in Stenosis Detection of Coronary Arteries. Expert Syst. Appl. 2023, 228, 120234.
- Zhang, Y.; Luo, G.; Wang, W.; Cao, S.; Dong, S.; Yu, D.; Wang, X.; Wang, K. TTN: Topological Transformer Network for Automated Coronary Artery Branch Labeling in Cardiac CT Angiography. IEEE J. Transl. Eng. Health Med. 2023, 12, 129–139.
- Rao, S.; Li, Y.; Ramakrishnan, R.; Hassaine, A.; Canoy, D.; Cleland, J.; Lukasiewicz, T.; Salimi-Khorshidi, G.; Rahimi, K.; Shishir Rao, O. An Explainable Transformer-Based Deep Learning Model for the Prediction of Incident Heart Failure. IEEE J. Biomed. Health Inf. 2022, 26, 3362–3372.
- Wang, Y.; Zhang, W. A Dense RNN for Sequential Four-Chamber View Left Ventricle Wall Segmentation and Cardiac State Estimation. Front. Bioeng. Biotechnol. 2021, 9, 696227.
- Ding, C.; Wang, S.; Jin, X.; Wang, Z.; Wang, J. A Novel Transformer-Based ECG Dimensionality Reduction Stacked Auto-Encoders for Arrhythmia Beat Detection. Med. Phys. 2023, 50, 5897–5912.
- Ding, Y.; Xie, W.; Wong, K.K.L.; Liao, Z. DE-MRI Myocardial Fibrosis Segmentation and Classification Model Based on Multi-Scale Self-Supervision and Transformer. Comput. Methods Programs Biomed. 2022, 226, 107049.
- Hu, R.; Chen, J.; Zhou, L. A Transformer-Based Deep Neural Network for Arrhythmia Detection Using Continuous ECG Signals. Comput. Biol. Med. 2022, 144, 105325.
- Gaudilliere, P.L.; Sigurthorsdottir, H.; Aguet, C.; Van Zaen, J.; Lemay, M.; Delgado-Gonzalo, R. Generative Pre-Trained Transformer for Cardiac Abnormality Detection. Available online: https://physionet.org/content/mitdb/1.0.0/ (accessed on 20 February 2024).
- Lecesne, E.; Simon, A.; Garreau, M.; Barone-Rochette, G.; Fouard, C. Segmentation of Cardiac Infarction in Delayed-Enhancement MRI Using Probability Map and Transformers-Based Neural Networks. Comput. Methods Programs Biomed. 2023, 242, 107841.
- Ahmadi, N.; Tsang, M.Y.; Gu, A.N.; Tsang, T.S.M.; Abolmaesumi, P. Transformer-Based Spatio-Temporal Analysis for Classification of Aortic Stenosis Severity from Echocardiography Cine Series. IEEE Trans. Med. Imaging 2024, 43, 366–376.
- Han, T.; Ai, D.; Li, X.; Fan, J.; Song, H.; Wang, Y.; Yang, J. Coronary Artery Stenosis Detection via Proposal-Shifted Spatial-Temporal Transformer in X-Ray Angiography. Comput. Biol. Med. 2023, 153, 106546.
- Ning, Y.; Zhang, S.; Xi, X.; Guo, J.; Liu, P.; Zhang, C. CAC-EMVT: Efficient Coronary Artery Calcium Segmentation with Multi-Scale Vision Transformers. In Proceedings of the 2021 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Houston, TX, USA, 9–12 December 2021; pp. 1462–1467.
- Deng, K.; Meng, Y.; Gao, D.; Bridge, J.; Shen, Y.; Lip, G.; Zhao, Y.; Zheng, Y. TransBridge: A Lightweight Transformer for Left Ventricle Segmentation in Echocardiography. In Simplifying Medical Ultrasound; Noble, J.A., Aylward, S., Grimwood, A., Min, Z., Lee, S.-L., Hu, Y., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 63–72.
- Alkhodari, M.; Kamarul Azman, S.; Hadjileontiadis, L.J.; Khandoker, A.H. Ensemble Transformer-Based Neural Networks Detect Heart Murmur in Phonocardiogram Recordings. In Proceedings of the 2022 Computing in Cardiology (CinC), Tampere, Finland, 4–7 September 2022.
- Meng, L.; Tan, W.; Ma, J.; Wang, R.; Yin, X.; Zhang, Y. Enhancing Dynamic ECG Heartbeat Classification with Lightweight Transformer Model. Artif. Intell. Med. 2022, 124, 102236.
- Chu, M.; Wu, P.; Li, G.; Yang, W.; Gutiérrez-Chico, J.L.; Tu, S. Advances in Diagnosis, Therapy, and Prognosis of Coronary Artery Disease Powered by Deep Learning Algorithms. JACC Asia 2023, 3, 1–14.
- Feng, Y.; Geng, S.; Chu, J.; Fu, Z.; Hong, S. Building and Training a Deep Spiking Neural Network for ECG Classification. Biomed. Signal Process. Control 2022, 77, 103749.
- Yan, Z.; Zhou, J.; Wong, W.F. Energy Efficient ECG Classification with Spiking Neural Network. Biomed. Signal Process. Control 2021, 63, 102170.
- Kovács, P.; Samiee, K. Arrhythmia Detection Using Spiking Variable Projection Neural Networks. In Proceedings of the 2022 Computing in Cardiology (CinC), Tampere, Finland, 4–7 September 2022; Volume 49.
- Singhal, S.; Kumar, M. GSMD-SRST: Group Sparse Mode Decomposition and Superlet Transform Based Technique for Multi-Level Classification of Cardiac Arrhythmia. IEEE Sens. J. 2024.
- Kiladze, M.R.; Lyakhova, U.A.; Lyakhov, P.A.; Nagornov, N.N.; Vahabi, M. Multimodal Neural Network for Recognition of Cardiac Arrhythmias Based on 12-Load Electrocardiogram Signals. IEEE Access 2023, 11, 133744–133754.
- Li, Z.; Calvet, L.E. Extraction of ECG Features with Spiking Neurons for Decreased Power Consumption in Embedded Devices Extraction of ECG Features with Spiking Neurons for Decreased Power Consumption in Embedded Devices Extraction of ECG Features with Spiking Neurons for Decreased Power Consumption in Embedded Devices. In Proceedings of the 2023 19th International Conference on Synthesis, Modeling, Analysis and Simulation Methods and Applications to Circuit Design (SMACD), Funchal, Portugal, 3–5 July 2023.
- Revathi, T.K.; Sathiyabhama, B.; Sankar, S. Diagnosing Cardio Vascular Disease (CVD) Using Generative Adversarial Network (GAN) in Retinal Fundus Images. Ann. Rom. Soc. Cell Biol. 2021, 25, 2563–2572. Available online: http://annalsofrscb.ro (accessed on 20 February 2024).
- Chen, J.; Yang, G.; Khan, H.R.; Zhang, H.; Zhang, Y.; Zhao, S.; Mohiaddin, R.H.; Wong, T.; Firmin, D.N.; Keegan, J. JAS-GAN: Generative Adversarial Network Based Joint Atrium and Scar Segmentations on Unbalanced Atrial Targets. IEEE J. Biomed. Health Inf. 2021, 26, 103–114.
- Zhang, Y.; Feng, J.; Guo, X.; Ren, Y. Comparative Analysis of U-Net and TLMDB GAN for the Cardiovascular Segmentation of the Ventricles in the Heart. Comput. Methods Programs Biomed. 2022, 215, 106614.
- Decourt, C.; Duong, L. Semi-Supervised Generative Adversarial Networks for the Segmentation of the Left Ventricle in Pediatric MRI. Comput. Biol. Med. 2020, 123, 103884.
- Diller, G.P.; Vahle, J.; Radke, R.; Vidal, M.L.B.; Fischer, A.J.; Bauer, U.M.M.; Sarikouch, S.; Berger, F.; Beerbaum, P.; Baumgartner, H.; et al. Utility of Deep Learning Networks for the Generation of Artificial Cardiac Magnetic Resonance Images in Congenital Heart Disease. BMC Med. Imaging 2020, 20, 113.
- Rizwan, I.; Haque, I.; Neubert, J. Deep Learning Approaches to Biomedical Image Segmentation. Inf. Med. Unlocked 2020, 18, 100297.
- Van Lieshout, F.E.; Klein, R.C.; Kolk, M.Z.; Van Geijtenbeek, K.; Vos, R.; Ruiperez-Campillo, S.; Feng, R.; Deb, B.; Ganesan, P.; Knops, R.; et al. Deep Learning for Ventricular Arrhythmia Prediction Using Fibrosis Segmentations on Cardiac MRI Data. In Proceedings of the 2022 Computing in Cardiology (CinC), Tampere, Finland, 4–7 September 2022.
- Liu, X.; He, L.; Yan, J.; Huang, Y.; Wang, Y.; Lin, C.; Huang, Y.; Liu, X. A Neural Network for High-Precise and Well-Interpretable Electrocardiogram Classification. bioRxiv 2023.
- Lu, P.; Bai, W.; Rueckert, D.; Noble, J.A. Modelling Cardiac Motion via Spatio-Temporal Graph Convolutional Networks to Boost the Diagnosis of Heart Conditions. In Statistical Atlases and Computational Models of the Heart; Springer: Berlin/Heidelberg, Germany, 2020.
- Yang, H.; Zhen, X.; Chi, Y.; Zhang, L.; Hua, X.-S. CPR-GCN: Conditional Partial-Residual Graph Convolutional Network in Automated Anatomical Labeling of Coronary Arteries. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020.
- Huang, F.; Lian, J.; Ng, K.-S.; Shih, K.; Vardhanabhuti, V.; Huang, F.; Lian, J.; Ng, K.-S.; Shih, K.; Vardhanabhuti, V. Citation: Predicting CT-Based Coronary Artery Disease Using Vascular Biomarkers Derived from Fundus Photographs with a Graph Convolutional Neural Network. Diagnostics 2022, 12, 1390.
- Gao, R.; Hou, Z.; Li, J.; Han, H.; Lu, B.; Zhou, S.K. Joint Coronary Centerline Extraction And Lumen Segmentation From Ccta Using Cnntracker And Vascular Graph Convolutional Network. In Proceedings of the 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), Nice, France, 13–16 April 2021; pp. 1897–1901.
- Chakravarty, A.; Sarkar, T.; Ghosh, N.; Sethuraman, R.; Sheet, D. Learning Decision Ensemble Using a Graph Neural Network for Comorbidity Aware Chest Radiograph Screening. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 1234–1237.
- Reddy, G.T.; Praveen, M.; Reddy, K.; Lakshmanna, K.; Rajput, D.S.; Kaluri, R.; Gautam, S. Hybrid Genetic Algorithm and a Fuzzy Logic Classifier for Heart Disease Diagnosis. Evol. Intell. 2020, 13, 185–196.
- Priyanka; Baranwal, N.; Singh, K.N.; Singh, A.K. YOLO-Based ROI Selection for Joint Encryption and Compression of Medical Images with Reconstruction through Super-Resolution Network. Future Gener. Comput. Syst. 2024, 150, 1–9.
- Wang, C.-Y.; Bochkovskiy, A.; Liao, H.-Y.M. YOLOv7: Trainable Bag-of-Freebies Sets New State-of-the-Art for Real-Time Object Detectors. arXiv 2022, arXiv:2207.02696.
- Zhuang, Z.; Jin, P.; Joseph Raj, A.N.; Yuan, Y.; Zhuang, S. Automatic Segmentation of Left Ventricle in Echocardiography Based on YOLOv3 Model to Achieve Constraint and Positioning. Comput. Math. Methods Med. 2021, 2021, 3772129.
- Alamelu, V.; Thilagamani, S. Lion Based Butterfly Optimization with Improved YOLO-v4 for Heart Disease Prediction Using IoMT. Inf. Technol. Control 2022, 51, 692–703.
- Smirnov, D.; Pikunov, A.; Syunyaev, R.; Deviatiiarov, R.; Gusev, O.; Aras, K.; Gams, A.; Koppel, A.; Efimov, I.R. Correction: Genetic Algorithm-Based Personalized Models of Human Cardiac Action Potential. PLoS ONE 2020, 15, e0231695.
- Kanwal, S.; Rashid, J.; Nisar, M.W.; Kim, J.; Hussain, A. An Effective Classification Algorithm for Heart Disease Prediction with Genetic Algorithm for Feature Selection. In Proceedings of the 2021 Mohammad Ali Jinnah University International Conference on Computing (MAJICC), Karachi, Pakistan, 15–17 July 2021.
- Alizadehsani, R.; Roshanzamir, M.; Abdar, M.; Beykikhoshk, A.; Khosravi, A.; Nahavandi, S.; Plawiak, P.; Tan, R.S.; Acharya, U.R. Hybrid Genetic-Discretized Algorithm to Handle Data Uncertainty in Diagnosing Stenosis of Coronary Arteries. Expert Syst. 2020, 39, e12573.
- Badano, L.P.; Keller, D.M.; Muraru, D.; Torlasco, C.; Parati, G. Artificial Intelligence and Cardiovascular Imaging: A Win-Win Combination. Anatol. J. Cardiol. 2020, 24, 214–223.
- Souza Filho, E.M.; Fernandes, F.A.; Pereira, N.C.A.; Mesquita, C.T.; Gismondi, R.A. Ethics, Artificial Intelligence and Cardiology. Arq. Bras. De Cardiol. 2020, 115, 579–583.
- Mathur, P.; Srivastava, S.; Xu, X.; Mehta, J.L. Artificial Intelligence, Machine Learning, and Cardiovascular Disease. Clin. Med. Insights Cardiol. 2020, 14, 1179546820927404.
- Miller, D.D. Machine Intelligence in Cardiovascular Medicine. Cardiol. Rev. 2020, 28, 53–64.
- Swathy, M.; Saruladha, K. A Comparative Study of Classification and Prediction of Cardio-Vascular Diseases (CVD) Using Machine Learning and Deep Learning Techniques. ICT Express 2021, 8, 109–116.
- Gao, Z.; Wang, L.; Soroushmehr, R.; Wood, A.; Gryak, J.; Nallamothu, B.; Najarian, K. Vessel Segmentation for X-Ray Coronary Angiography Using Ensemble Methods with Deep Learning and Filter-Based Features. BMC Med. Imaging 2022, 22, 10.
- Galea, R.R.; Diosan, L.; Andreica, A.; Popa, L.; Manole, S.; Bálint, Z. Region-of-Interest-Based Cardiac Image Segmentation with Deep Learning. Appl. Sci. 2021, 11, 1965.
- Tandon, A.; Mohan, N.; Jensen, C.; Burkhardt, B.E.U.; Gooty, V.; Castellanos, D.A.; McKenzie, P.L.; Zahr, R.A.; Bhattaru, A.; Abdulkarim, M.; et al. Retraining Convolutional Neural Networks for Specialized Cardiovascular Imaging Tasks: Lessons from Tetralogy of Fallot. Pediatr. Cardiol. 2021, 42, 578–589.
- Stough, J.V.; Raghunath, S.; Zhang, X.; Pfeifer, J.M.; Fornwalt, B.K.; Haggerty, C.M. Left Ventricular and Atrial Segmentation of 2D Echocardiography with Convolutional Neural Networks. In Proceedings of the Medical Imaging 2020: Image Processing, Houston, TX, USA, 10 March 2020.
- Sander, J.; de Vos, B.D.; Išgum, I. Automatic Segmentation with Detection of Local Segmentation Failures in Cardiac MRI. Sci. Rep. 2020, 10, 21769.
- Lin, A.; Chen, B.; Xu, J.; Zhang, Z.; Lu, G. DS-TransUNet: Dual Swin Transformer U-Net for Medical Image Segmentation. IEEE Trans. Instrum. Meas. 2022, 71, 4005615.
- Koresh, H.J.D.; Chacko, S.; Periyanayagi, M. A Modified Capsule Network Algorithm for Oct Corneal Image Segmentation. Pattern Recognit. Lett. 2021, 143, 104–112.
- Khan, M.Z.; Gajendran, M.K.; Lee, Y.; Khan, M.A. Deep Neural Architectures for Medical Image Semantic Segmentation: Review. IEEE Access 2021, 9, 83002–83024.
- Lu, X.H.; Liu, A.; Fuh, S.C.; Lian, Y.; Guo, L.; Yang, Y.; Marelli, A.; Li, Y. Recurrent Disease Progression Networks for Modelling Risk Trajectory of Heart Failure. PLoS ONE 2021, 16, e0245177.
- Fu, Q.; Dong, H. An Ensemble Unsupervised Spiking Neural Network for Objective Recognition. Neurocomputing 2021, 419, 47–58.
- Rana, A.; Kim, K.K. A Novel Spiking Neural Network for ECG Signal Classification. J. Sens. Sci. Technol. 2021, 30, 20–24.
- Kim, Y.; Panda, P. Visual Explanations from Spiking Neural Networks Using Inter-Spike Intervals. Sci. Rep. 2021, 11, 19037.
- Kazeminia, S.; Baur, C.; Kuijper, A.; van Ginneken, B.; Navab, N.; Albarqouni, S.; Mukhopadhyay, A. GANs for Medical Image Analysis. Artif. Intell. Med. 2020, 109, 101938.
- Olender, M.L.; Nezami, F.R.; Athanasiou, L.S.; De La Torre Hernández, J.M.; Edelman, E.R. Translational Challenges for Synthetic Imaging in Cardiology. Eur. Heart J. Digit. Health 2021, 2, 559–560.
- Wieneke, H.; Voigt, I. Principles of Artificial Intelligence and Its Application in Cardiovascular Medicine. Clin. Cardiol. 2023, 47, e24148.
- Hasan Rafi, T.; Shubair, R.M.; Farhan, F.; Hoque, Z.; Mohd Quayyum, F. Recent Advances in Computer-Aided Medical Diagnosis Using Machine Learning Algorithms with Optimization Techniques. IEEE Access 2021, 9, 137847–137868.
- Carion, N.; Massa, F.; Synnaeve, G.; Usunier, N.; Kirillov, A.; Zagoruyko, S. End-to-End Object Detection with Transformers. Available online: https://github.com/facebookresearch/detr (accessed on 20 February 2024).
- Huang, J.; Ren, L.; Feng, L.; Yang, F.; Yang, L.; Yan, K. AI Empowered Virtual Reality Integrated Systems for Sleep Stage Classification and Quality Enhancement. IEEE Trans. Neural. Syst. Rehabil. Eng. 2022, 30, 1494–1503.
- Jannatul Ferdous, G.; Akhter Sathi, K.; Hossain, A.; Moshiul Hoque, M.; Member, S.; Ali Akber Dewan, M. LCDEiT: A Linear Complexity Data-Efficient Image Transformer for MRI Brain Tumor Classification. IEEE Access 2017, 11, 20337–20350.
- Beer, K.; Bondarenko, D.; Farrelly, T.; Osborne, T.J.; Salzmann, R.; Scheiermann, D.; Wolf, R. Training Deep Quantum Neural Networks. Nat. Commun. 2020, 11, 808.
- Landman, J.; Mathur, N.; Li, Y.Y.; Strahm, M.; Kazdaghli, S.; Prakash, A.; Kerenidis, I. Quantum Methods for Neural Networks and Application to Medical Image Classification. Quantum 2022, 6, 881.
- Shahwar, T.; Zafar, J.; Almogren, A.; Zafar, H.; Rehman, A.U.; Shafiq, M.; Hamam, H. Automated Detection of Alzheimer’s via Hybrid Classical Quantum Neural Networks. Electronics 2022, 11, 721.
- Ovalle-Magallanes, E.; Avina-Cervantes, J.G.; Cruz-Aceves, I.; Ruiz-Pinales, J. Hybrid Classical–Quantum Convolutional Neural Network for Stenosis Detection in X-Ray Coronary Angiography. Expert Syst. Appl. 2022, 189, 116112.
- Kumar, A.; Choudhary, A.; Tiwari, A.; James, C.; Kumar, H.; Kumar Arora, P.; Akhtar Khan, S. An Investigation on Wear Characteristics of Additive Manufacturing Materials. Mater. Today Proc. 2021, 47, 3654–3660.
- Belli, C.; Sagingalieva, A.; Kordzanganeh, M.; Kenbayev, N.; Kosichkina, D.; Tomashuk, T.; Melnikov, A. Hybrid Quantum Neural Network For Drug Response Prediction. Cancers 2023, 15, 2705.
- Pregowska, A.; Perkins, M. Artificial Intelligence in Medical Education: Technology and Ethical Risk. Available online: https://ssrn.com/abstract=4643763 (accessed on 20 February 2024).
- Rastogi, D.; Johri, P.; Tiwari, V.; Elngar, A.A. Multi-Class Classification of Brain Tumour Magnetic Resonance Images Using Multi-Branch Network with Inception Block and Five-Fold Cross Validation Deep Learning Framework. Biomed. Signal Process. Control 2024, 88, 105602.
