Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Oncology has emerged as a crucial field of study in the domain of medicine. Computed tomography has gained widespread adoption as a radiological modality for the identification and characterisation of pathologies, particularly in oncology, enabling precise identification of affected organs and tissues. However, achieving accurate liver segmentation in computed tomography scans remains a challenge due to the presence of artefacts and the varying densities of soft tissues and adjacent organs.
- artificial intelligence
- computed tomography
- hepatic pathologies
- liver segmentation
1. Introduction
As one of the most important organs in the digestive system, the liver performs critical functions such as breaking down nutrients, producing bile and eliminating toxic substances. However, liver-related diseases, particularly oncological diseases, pose health risks, and liver cancer is a leading cause of cancer-related mortality worldwide [1][2]. Computed tomography (CT) has become an integral part of diagnosis, treatment planning and monitoring the progress of oncological diseases [3][4], providing detailed cross-sectional images for accurate visualisation of internal structures, including liver tumours [5][6].
With the advancement of technology and artificial intelligence (AI) in medicine, there is a growing need to optimise the identification of oncological diseases [7]. Medical image segmentation is emerging as a fundamental step in the pipeline [8][9][10][11]. Liver segmentation in CT scans has emerged as a critical area, requiring accurate identification and delimitation of the liver region for treatment planning and progress monitoring, as well as for early detection of liver lesions and metastases to other organs [12][13][14]. However, accurate liver segmentation on CT scans is challenging due to factors such as artefacts, varying soft tissue densities and the complexity caused by adjacent organ proximity [15][16].
2. Historical Overview
The oldest work found to tackle liver segmentation is the one by Bae et al. (1993) [17], presenting a similar sequential image-by-image segmentation technique using a reference image, where the liver occupies a significant portion of the abdomen cross-section. Image processing techniques, including grey-level thresholding, Gaussian smoothing and connectivity tracking, are employed to extract the liver boundaries. The resulting boundaries are then smoothed using mathematical morphology techniques and B-splines. This study focuses on a living-donor liver transplant program, and the computer-determined boundaries are compared with those drawn by a radiologist, showing agreement within 10% of the calculated areas.
Gao et al. (1996) [18] focus on facilitating 3D visualisations for surgical planning. Their method employs a global histogram analysis, morphologic operations and a parametrically deformable contour model to delineate the liver boundary. Ten cases were used to validate the approach and promising results were found with minimal operator intervention required.
Soler et al. (1997) [19] propose an automatic method for segmenting the portal vein, with the primary objective of achieving accurate segmentation with detailed branching and topological information, facilitating the localisation of liver tumours concerning Couinaud’s anatomical segmentation. This approach involves the initial detection of liver contours using 3D deformable models, followed by limiting the CT images to a liver mask containing hepatic tissue, vascular trees and potential tumours. Classification of anatomical structures is performed using Gaussian curves fitted to an intensity histogram. The vascular trees and tumours are segmented through a hysteresis thresholding technique based on a distance map, considering the Gaussian parameters. An isotropic image is obtained through shape-based interpolation and the portal vein is reconstructed using skeletonization, eliminating short branches and correcting errors. Results demonstrate that the algorithm automatically extracts the first three main bifurcations of the portal vein, comparable to manual segmentation.
Yoo et al. (2000) [20] focus on the use of pixel ratios. By analysing the grey value range of a normal liver in CT images, a binary image is generated and then processed into four mesh images based on hole ratios to eliminate noise. A template representing the general outline of the liver is generated from the union image of these mesh images and subtracted from the binary image to accurately represent the organ boundary. The pixel ratio, which takes into account the distribution of organ pixels, was used to discriminate between the organ and noise, especially in cases where organs have similar grey value ranges. The proposed method reduced the processing time compared to existing methods and was validated against manual segmentation by medical experts.
Pan and Dawant (2001) [21] introduce a level-set approach, which addresses the challenge of defining appropriate speed functions for contour propagation. A speed function is proposed to stop the propagation of the contour at organ boundaries with weak edges by incorporating the accumulative speed based on the path of the contour, enhancing the robustness of segmentation in noisy images. The method also leverages a priori anatomical information to improve the accuracy. Tested on five CT datasets, including cases with abnormal livers, tise method demonstrates good agreement with manual delineations.
Saitoh et al. (2002) [22] present an automated method for segmenting the liver region from the third phase of abdominal CT scans. Their approach involves the extraction of blood vessels using a threshold, followed by morphological dilation to define an approximate liver region useful for the removal of adjacent organs. The final liver region is then extracted using a threshold. The method is thus based on mathematical morphology and thresholding techniques, using the unique characteristics of blood vessels to functionally identify the liver region. The experiments performed on eight CT datasets show a good agreement between the automatically and manually detected liver regions.
Masumoto et al. (2003) [23] use multislice CT images. Their method uses two time-varying images acquired during the contrast medium circulation phase, highlighting the liver region through CT value changes. The proposed scheme involves generating a liver likelihood image by analysing CT value changes and subsequently extracting the liver region while considering the geometric characteristics of blood vessels and tumours. The evaluation, based on receiver operating characteristic (ROC) analyses, demonstrates the superiority of the proposed method over other approaches, especially when using information from both phases.
The scheme proposed by Lim et al. (2004) [24] uses an ROI approach to optimise computational efficiency. Morphological filters, incorporating a priori knowledge of liver location and intensity, detect the initial boundary. The algorithm then generates a gradient image using the weighted initial boundary and employs an immersion-based watershed algorithm for segmentation. Post-processing includes region merging based on statistical information to refine the segmentation.
Liu et al. (2005) [25] present a gradient vector flow (GVF) snake-based method for the semi-automatic segmentation of liver volumes in contrast-enhanced CT images. The algorithm follows a stepwise approach, starting with the computation of an initial edge map using the Canny edge detector and the estimation of a liver template. The edge map is then modified to suppress edges within the liver using the liver template, and a concavity removal algorithm is applied to refine the liver boundary. The GVF field is computed based on the modified edge map, and the initial liver contour is determined by considering the candidate initial contour and the computed GVF field. The final liver contour is obtained by deforming the initial contour using the snake. The method was evaluated on 20 contrast-enhanced volumetric liver images, and the results were compared with a radiologist’s manual delineation. The median difference ratio between the computer-generated results and manual results is 5.3%, with a range of 2.9% to 7.6%.
A three-stage approach is used by Lim et al. (2006) [26]. The first stage involves image simplification as preprocessing, where an ROI is identified and thresholds are determined using multilevel thresholding. The second stage detects a search range using multiscale morphological filtering, region labelling, and partition clustering. The third stage uses a contour-based segmentation approach with a labelling-based search algorithm to refine the initial liver boundary. The effectiveness of the algorithm is demonstrated through experimental results on contrast-enhanced abdominal CT images, with an average segmentation accuracy of 96%. Volume measurement is performed based on the segmented liver regions, with an average error rate of 3%.
Beichel et al. (2007) [27] introduce a two-step process. First, initial segmentation is performed using graph cuts, overcoming challenges such as the high variability in liver shape and grey-value appearance. Second, an interactive refinement step is introduced, allowing users to correct segmentation errors in a 3D environment. The refinement is facilitated by a hybrid desktop/virtual reality (VR) user interface. This approach is demonstrated on ten contrast-enhanced liver CT scans, demonstrating robustness to variations in patient data. The results also indicate an improved segmentation quality with low interaction times.
Massoptier and Casciaro (2008) [28] present a fully automated method that uses a statistical-model-based approach to distinguish liver tissue from other abdominal organs. An active contour technique using gradient vector flow is used for smoother segmentation of the liver surface. Automatic classification is performed to isolate hepatic lesions from liver parenchyma. The method was evaluated on 21 datasets and demonstrated robust and efficient liver and lesion segmentations close to the ground truth, with an average processing time of 11.4 s per 512 × 512 pixel slice. The volume overlap for liver surface segmentation is 94.2%, and the accuracy is 3.7 mm. Tumour detection achieved a sensitivity and specificity of 82.6% and 87.5%, respectively.
Heimann et al. (2009) [29] focus on the comparison and evaluation of different methods. The image data, acquired from different CT scanners, consisted of contrast-dye-enhanced scans showing pathological conditions like tumours and cysts. Radiology experts manually delineated the liver contours in transversal slices to create reference segmentations. A total of 40 images were divided into training and test sets for algorithm evaluation. Evaluation measures included volumetric overlap, relative volume difference, and surface distances. Fully automated and interactive segmentation methods were employed, with the former showing discernible performance differences. The best-performing automated approaches used statistical shape models. Interactive methods achieved higher scores with more user interaction. A combined approach using majority voting from the best-performing methods outperformed individual automated and interactive results.
A three-step procedure is outlined by Akram et al. (2010) [30]. Firstly, a pre-processing step involves converting the image to greyscale and applying a 3 × 3 median filter to reduce noise. The second step focuses on liver segmentation, with a global threshold and morphological operations to obtain the final segmented liver region. Finally, post-processing steps include adaptive histogram equalisation, Gaussian smoothing, and grey-level transformations to enhance the segmented liver region. Experimental tests on 100 CT images demonstrate the accuracy of the proposed method by comparing automated segmentation results with images manually segmented by hepatologists and oncologists.
The approach of Oliveira et al. (2011) [31] involves a sequence of four steps. First, the liver is segmented using level sets with parameters optimised by a genetic algorithm (GA). A Gaussian fit is employed to define the speed image for level set propagation. Secondly, vessels and nodules are segmented using a Gaussian mixture model, focusing on adipose nodules. A region-growing method with information from the Gaussian model is applied. Thirdly, vessels are classified into portal veins or hepatic veins using a vein tracking method. Finally, a geometric approach based on the identified veins is used to segment the liver into different Couinaud regions. Liver segmentation is based on the assumption that the liver parenchyma homogeneity and veins are mainly inside the liver. The parameters are estimated using a GA, and the fitness evaluation involves comparing the segmentation with a reference using five disparity metrics. The proposed method shows good performance, ranking among the top methods in the MICCAI-SLiver07 conference evaluation.
The method developed by Linguraru et al. (2012) [32] uses a robust parametrisation of 3D surfaces for point-to-point correspondence, overcoming challenges such as inconsistent contrast enhancements and imaging artefacts. A shape descriptor that is invariant under rotation and scale is used to compare the local shape features of organs. Initial liver segmentation is refined using a shape-driven geodesic active contour, and hepatic tumours are detected and segmented using graph cuts and support vector machines (SVMs). This technique is evaluated on a dataset of 101 CT scans and shows improvements in the liver segmentation accuracy, particularly in cases with large tumours and segmentation errors. Furthermore, the method identifies liver tumours with a low rate of false positives.
Li et al. (2013) [33] discuss a method that makes use of fuzzy clustering and level set techniques. The Fuzzy C-Means (FCM) clustering algorithm is employed, which assigns pixels to different categories based on fuzzy memberships, considering both the grey level intensity and spatial information. The FCM algorithm is iteratively optimised by minimising a cost function, allowing the fuzziness of the resulting partition. To overcome the limitations of standard FCM, a spatial FCM algorithm is introduced that incorporates spatial information into fuzzy membership functions. This paper also introduces the level set method, a continuous deformable model for segmentation. Distance-Regularized Level Set Evolution (DRLSE) is proposed to address reinitialisation issues and improve the efficiency. The proposed method is evaluated using accuracy, sensitivity, and specificity metrics and demonstrates a high performance in liver segmentation, especially in cases with unclear boundaries. A comprehensive review of abdominal image segmentation using soft and hard computing approaches is provided in [34].
Platero et al. (2014) [35] integrate a multi-atlas segmentation approach with graph cuts. Their method includes several steps: (1) obtaining an initial solution using low-level operations to define the ROI around the liver; (2) constructing a fast probabilistic atlas for the ROI and computing a coarse binary segmentation using segmentation-affine registration; (3) ranking the atlases based on segmentation similarity and propagating selected atlases to the target image; (4) improving the segmentation accuracy through label fusion, minimising the discrete energy function; and (5) evaluating the approach using a public liver segmentation database. The experimental results show a high accuracy, competitive with human expert segmentation.
Artificial Bee Colony (ABC) optimisation is used by Mostafa et al. (2015) [36]. Their algorithm use ABC optimisation to cluster different intensity values in abdominal CT images, followed by mathematical morphological operations to manipulate and separate the clusters. This process eliminates small and thin regions, such as flesh regions or organ edges. The extracted regions form an initial estimate of the liver area, which is further enhanced using a region-growing technique. The proposed approach demonstrates a segmentation accuracy of 93.73% on a test dataset of 38 CT images, taken in the pre-contrast phase.
A 3D deeply supervised network (DSN) is introduced by Dou et al. (2016) [37]. The proposed architecture consists of 11 layers, including 6 convolutional layers, 2 max-pooling layers, 2 deconvolution layers, and 1 softmax layer. The network is designed in a 3D format to effectively capture spatial information. The 3D DSN employs deep supervision via additional deconvolutional layers to counteract vanishing gradients, thus improving the training process. The learning objective is to minimise per-voxel-wise binary classification errors, with deep supervision injected at specific layers. The MICCAI-SLiver07 dataset is used for evaluation, demonstrating that the 3D DSN has a faster convergence and lower errors when compared to traditional 3D convolutional neuronal networks (CNNs).
Christ et al. (2017) [14] propose a cascaded fully CNN (CFCN) on CT slices that sequentially segments the liver and lesions. First, various preprocessing steps, including Hounsfield unit windowing and contrast enhancement, are applied. Then, the cascaded approach involving two U-Net architectures is used for liver and lesion segmentation. Finally, 3D conditional random fields (CRFs) are used to refine the segmentation results. Generalisation and scalability to different modalities and real-life datasets, including a diffusion-weighted magnetic resonance imaging (MRI) dataset and a large multi-centre CT dataset, are shown.
Hiraman (2018) [38] presents a slice alignment method that addresses the challenges through optimal threshold selection, skeletonization, and enhanced correlation coefficient (ECC) alignment. Next, a CNN-based liver region of interest detection method is proposed to classify 2D slices for focused processing.
The study presented by Wang et al. (2019) [39] investigates the application of a generalised CNN for automated liver segmentation and biometry using cross-sectional data from abdominal CT and MRI scans. Their retrospective study included a sample of 563 abdominal scans from 530 adults, covering different imaging modalities. The CNN was initially trained on 300 unenhanced multiecho 2D SPGR MRI sets and then subjected to transfer learning for generalisation across different imaging methods. The accuracy of the CNN was evaluated using internal and external validation datasets. This study also investigates the impact of training data size on the segmentation accuracy and explores the feasibility of using automated liver segmentation for volumetry and hepatic PDFF quantification.
Almotairi et al. (2020) [40] explore the application of the SegNet architecture. The proposed modified SegNet model uses the VGG-16 network as an encoder. Tests were performed on a standard dataset for liver CT scans (3D-IRCADb01 [41]), achieving a tumour accuracy of up to 99.9% in the training phase and 86% for tumour identification.
Ayalew et al. (2021) [42] present a modified U-Net architecture and introduce a new class balancing method. To address the class imbalance between the liver and tumours, a weighting factor is applied and slices without a tumour are removed during data preparation. The U-Net-based network architecture includes batch normalisation, dropout layers, and filter size reduction. Training involves tuning hyperparameters, such as the learning rate and batch size. The datasets used are derived from the 3D-IRCADb01 [41] and LiTS [43] databases and the results achieve a Dice Similarity Coefficient (DSC) of 0.96 and 0.74, respectively. The algorithm also introduces a novel approach for direct tumour segmentation from abdominal CT scan images, with a comparable performance to existing two-step methods.
The study of Scicluna (2022) [44] is motivated by challenges such as the Combined Healthy Abdominal Organ Segmentation (CHAOS) Challenge [45], which focuses on healthy abdominal organs. The study focuses on replicating the v16pUNet1.1C model, which demonstrated a superior performance in Task 2 of the CHAOS Challenge. Results from the v16pUNet1.1C model are presented and compared with variations in the loss function and scaling transformation. The application of a 3D largest-connected-component filter is discussed, showing improvements in mean scores.
A deep semantic segmentation CNN is used by Ezzat et al. (2023) [46]. A three-stage architecture is proposed, including pre-processing with data augmentation, deep CNN training, and testing. The CNN-based semantic segmentation model is shown to be robust, achieving a test accuracy of 98.8%. The approach does not require user input, making it accessible to non-experts.
Shao et al. (2024) [47] present the Attention Connect Network (AC-Net) for liver tumour segmentation in CT and MRI images. The AC-Net consists of two main modules: an axial attention module (AAM) and a Vision Transformer module (VTM). The AAM uses an axial attention mechanism to merge features of matching dimensions, maximising the use of spatial features extracted by a CNN. The VTM processes high-level semantic features extracted by the CNN using a methodology similar to Vision Transformers (ViTs) [48]. The network achieves a DSC of 0.90, a Jaccard coefficient (JC) of 0.82, a recall of 0.92, a precision of 0.89, a Hausdorff distance (HD) of 11.96, and an average symmetric surface distance (ASSD) of 4.59.
3. Public Dataset Analysis
The most common public datasets used in studies of liver segmentation on CT scans include 3D-IRCADb01, LiTS17, and MICCAI-SLiver07 (Figure 1). According to a dataset comparison provided by Al-Saeed et al. [49] (shown in Table 1), it is possible to identify several key differences that may have implications for data processing and analysis, such as different formats and differences in resolution between datasets which may require different approaches to processing and interpretation.
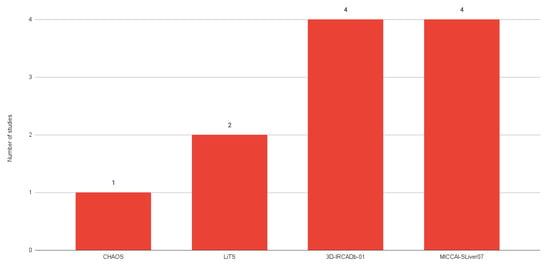
Figure 1. Number of studies using each public dataset.
Table 1. Characteristics of each dataset used by the main analysed studies (based on [49]).
4. Impact of the Adoption of Neuronal-Network-Based Methods
The growth of neuronal-network-based approaches has led to remarkable progress in liver segmentation, particularly with respect to CT scans. These models have led to a new era of accuracy and efficiency, significantly outperforming traditional methods [51][52]. This improved accuracy has become critical in the field of medical imaging, where the correct interpretation of CT scans can directly affect the diagnosis and treatment plans of patients. Furthermore, the efficiency of these neuronal network models translates into faster processing times, allowing for more agile decision making in clinical settings.
Another positive aspect of neuronal networks in liver segmentation is their ability to cope with the complexity of the liver anatomy. Neuronal networks, with their systematic and complex pattern recognition capabilities, are able to navigate these anatomical variations. As a result, they are better able to deal with the variety of appearances that liver tissue can have on CT scans. This ability to handle complex datasets ensures that neuronal networks can provide consistent and accurate segmentation in a wide range of cases.
5. Comparison between 2D and 3D Methods for Liver Segmentation
-
Importance of Choosing between 2D and 3D Methods
- -
-
In medical imaging, and in particular liver segmentation, the choice between slice-based 2D and volume-based 3D segmentation methods is crucial. This decision is highly dependent on the anatomical structure of the liver. Given the complex, three-dimensional nature of the liver, 3D segmentation techniques often prove to be the most appropriate choice [19][38]. These methods are inherently designed to understand and process the volumetric characteristics of the liver, which is a critical consideration for accurate segmentation results.
-
Two-Dimensional Segmentation Limitations
- -
-
Although 2D slice-based segmentation is widely used, it has limitations, particularly when it comes to dealing with complex organs such as the liver. The main challenge with 2D methods is their inability to fully capture all the regions of the liver. They involve working with individual slices, which can provide a fragmented understanding of the organ structure, but this fragmentation can lead to inconsistencies and errors when these individual slices are aggregated to form a complete image [39].
-
Three-Dimensional Segmentation Advantages
- -
-
In order to overcome the limitations of 2D segmentation, 3D segmentation has the ability to use more contextual information. Unlike 2D methods, which visualise the liver in individual slices, 3D techniques consider the organ as a whole, as they have the ability to ensure anatomical correctness by processing the liver as a single, continuous volume, avoiding errors that can arise from the aggregation of 2D slices [14][37]. In 2D segmentation, inconsistencies can occur when individual slices are combined, leading to inaccuracies in the representation of the liver anatomy. The holistic view provided by 3D segmentation results in more accurate segmentation, as it takes into account the spatial relationships and continuity between the different sections of the liver. The inclusion of this additional contextual information can potentially lead to segmentation results, especially in complex cases where the shape and size of the liver can vary considerably.
This entry is adapted from the peer-reviewed paper 10.3390/s24061752
References
- Francque, S. The Liver and the Cardiovascular System: Two of a Kind? J. Am. Heart Assoc. 2021, 10, e020286.
- Trefts, E.; Gannon, M.; Wasserman, D.H. The liver. Curr. Biol. 2017, 27, R1147–R1151.
- Mendes, B.; Domingues, I.; Silva, A.; Santos, J. Prostate Cancer Aggressiveness Prediction Using CT Images. Life 2021, 11, 1164.
- Pereira, G.; Domingues, I.; Martins, P.; Abreu, P.H.; Duarte, H.; Santos, J. Registration of CT with PET: A Comparison of Intensity-Based Approaches. In International Workshop on Combinatorial Image Analysis (IWCIA); Springer: Berlin/Heidelberg, Germany, 2018; pp. 134–149.
- Zhang, H.; Luo, K.; Deng, R.; Li, S.; Duan, S. Deep Learning-Based CT Imaging for the Diagnosis of Liver Tumor. Comput. Intell. Neurosci. 2022, 2022, 1–7.
- Vernuccio, F.; Cannella, R.; Bartolotta, T.V.; Galia, M.; Tang, A.; Brancatelli, G. Advances in liver US, CT, and MRI: Moving toward the future. Eur. Radiol. Exp. 2021, 5, 52.
- Domingues, I.; Cardoso, J.S. Using Bayesian surprise to detect calcifications in mammogram images. In Proceedings of the 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Chicago, IL, USA, 26–30 August 2014; pp. 1091–1094.
- Mendes, B.; Domingues, I.; Santos, J. Multi-class Semantic Segmentation for Prostate Cancer Radiotherapy Treatment Optimization. In Proceedings of the International Conference on Mathematical Analysis and Applications in Science and Engineering (ICMA2SC), Porto, Portugal, 27–29 June 2022.
- Bechar, M.E.A.; Settouti, N.; Domingues, I. Deep Learning vs. Super Pixel Classification for Breast Masses Segmentation. In Deep Learning for Biomedical Applications; CRC Press: Boca Raton, FL, USA, 2021; pp. 121–156.
- Oliveira, A.C.; Domingues, I.; Duarte, H.; Santos, J.; Abreu, P.H. Going Back to Basics on Volumetric Segmentation of the Lungs in CT: A Fully Image Processing Based Technique. In Iberian Conference on Pattern Recognition and Image Analysis (IbPRIA); Springer: Berlin/Heidelberg, Germany, 2019; Volume 11868 LNCS, pp. 322–334.
- Carbone, I.; Martins, P.; Teixeira, A.; Silva, A. A Vocal Tract Segmentation and Analysis over a European Portuguese MRI Database. Electrónica E Telecomunicações 2008, 4, 1050–1053.
- Zhou, L.Q.; Wang, J.Y.; Yu, S.Y.; Wu, G.G.; Wei, Q.; Deng, Y.B.; Wu, X.L.; Cui, X.W.; Dietrich, C.F. Artificial intelligence in medical imaging of the liver. World J. Gastroenterol. 2019, 25, 672–682.
- Chlebus, G.; Schenk, A.; Moltz, J.H.; van Ginneken, B.; Hahn, H.K.; Meine, H. Automatic liver tumor segmentation in CT with fully convolutional neural networks and object-based postprocessing. Sci. Rep. 2018, 8, 15497.
- Christ, P.F.; Ettlinger, F.; Grün, F.; Elshaera, M.E.A.; Lipkova, J.; Schlecht, S.; Ahmaddy, F.; Tatavarty, S.; Bickel, M.; Bilic, P.; et al. Automatic Liver and Tumor Segmentation of CT and MRI Volumes using Cascaded Fully Convolutional Neural Networks. arXiv 2017, arXiv:1702.05970.
- Ansari, M.Y.; Abdalla, A.; Ansari, M.Y.; Ansari, M.I.; Malluhi, B.; Mohanty, S.; Mishra, S.; Singh, S.S.; Abinahed, J.; Al-Ansari, A.; et al. Practical utility of liver segmentation methods in clinical surgeries and interventions. BMC Med. Imaging 2022, 22, 97.
- Le, D.C.; Chinnasarn, K.; Chansangrat, J.; Keeratibharat, N.; Horkaew, P. Semi-automatic liver segmentation based on probabilistic models and anatomical constraints. Sci. Rep. 2021, 11, 6106.
- Bae, K.T.; Giger, M.L.; Chen, C.T.; Kahn, C.E., Jr. Automatic segmentation of liver structure in CT images. Med. Phys. 1993, 20, 71–78.
- Gao, L.; Heath, D.G.; Kuszyk, B.S.; Fishman, E.K. Automatic liver segmentation technique for three-dimensional visualization of CT data. Radiology 1996, 201, 359–364.
- Soler, L.; Malandain, G.; Montagnat, J.; Delingette, H.; Ayache, N.; Clément, J.M.; Roy, C.; Russier, Y.; Tassetti, V.; Marescaux, J. Automatic Segmentation of Portal Vein in CT-Scans of the Liver. In Proceedings of the World Congress on Medical Physics and Biomedical Engineering (MPBE), Nice, France, 14–19 September 1997; p. 788.
- Yoo, S.W.; Cho, J.S.; Noh, S.M.; Shin, K.S.; Park, J.W. Advanced Liver Segmentation by Using Pixel Ratio in Abdominal CT Image. In Proceedings of the IEEK Conference, Pusan, Korea, 2000; The Institute of Electronics and Information Engineers: Seoul, Korea, 2000; pp. 39–42.
- Pan, S.; Dawant, B.M. Automatic 3D segmentation of the liver from abdominal CT images: A level-set approach. In Medical Imaging: Image Processing; SPIE: Bellingham, WA, USA, 2001; Volume 4322, pp. 128–138.
- Saitoh, T.; Tamura, Y.; Kaneko, T. Automatic segmentation of liver region through blood vessels on multi-phase CT. In Proceedings of the International Conference on Pattern Recognition, Quebec, ON, Canada, 11–15 August 2002; IEEE: Piscataway, NJ, USA, 2002; Volume 1, pp. 735–738.
- Masumoto, J.; Hori, M.; Sato, Y.; Murakami, T.; Johkoh, T.; Nakamura, H.; Tamura, S. Automated liver segmentation using multislice CT images. Syst. Comput. Jpn. 2003, 34, 71–82.
- Lim, S.J.; Jeong, Y.Y.; Lee, C.W.; Ho, Y.S. Automatic segmentation of the liver in CT images using the watershed algorithm based on morphological filtering. In Proceedings of the Medical Imaging: Image Processing, San Diego, CA, USA, 6–19 February 2004; SPIE: Bellingham, WA, USA, 2004; Volume 5370, pp. 1658–1666.
- Liu, F.; Zhao, B.; Kijewski, P.K.; Wang, L.; Schwartz, L.H. Liver segmentation for CT images using GVF snake. Med. Phys. 2005, 32, 3699–3706.
- Lim, S.J.; Jeong, Y.Y.; Ho, Y.S. Automatic liver segmentation for volume measurement in CT Images. J. Vis. Commun. Image Represent. 2006, 17, 860–875.
- Beichel, R.; Bauer, C.; Bornik, A.; Sorantin, E.; Bischof, H. Liver Segmentation in CT Data: A Segmentation Refinement Approach. In Segmentation in The Clinic: A Grand Challenge; 2007; pp. 235–245. Available online: https://graz.elsevierpure.com/en/publications/liver-segmentation-in-ct-data-a-segmentation-refinement-approach (accessed on 4 March 2024).
- Massoptier, L.; Casciaro, S. A new fully automatic and robust algorithm for fast segmentation of liver tissue and tumors from CT scans. Eur. Radiol. 2008, 18, 1658–1665.
- Heimann, T.; Van Ginneken, B.; Styner, M.A.; Arzhaeva, Y.; Aurich, V.; Bauer, C.; Beck, A.; Becker, C.; Beichel, R.; Bekes, G.; et al. Comparison and evaluation of methods for liver segmentation from CT datasets. IEEE Trans. Med Imaging 2009, 28, 1251–1265.
- Akram, M.U.; Khanum, A.; Iqbal, K. An automated system for liver CT enhancement and segmentation. Icgst J. Graph. Vis. Image Process. (ICGST-GVIP) 2010, 10, 17–22.
- Oliveira, D.A.; Feitosa, R.Q.; Correia, M.M. Segmentation of liver, its vessels and lesions from CT images for surgical planning. Biomed. Eng. Online 2011, 10, 1–23.
- Linguraru, M.G.; Richbourg, W.J.; Liu, J.; Watt, J.M.; Pamulapati, V.; Wang, S.; Summers, R.M. Tumor Burden Analysis on Computed Tomography by Automated Liver and Tumor Segmentation. IEEE Trans. Med. Imaging 2012, 31, 1965–1976.
- Li, X.; Luo, S.; Li, J. Liver Segmentation from CT Image Using Fuzzy Clustering and Level Set. J. Signal Inf. Process. 2013, 4, 36–42.
- Jena, B.; Krishna Nayak, G.; Saxena, S. Comprehensive Review of Abdominal Image Segmentation using Soft and Hard Computing Approaches. In Proceedings of the International Conference on Computer Science, Engineering and Applications (ICCSEA), Gunupur, India, 13–14 March 2020; pp. 1–5.
- Platero, C.; Tobar, M.C. A multiatlas segmentation using graph cuts with applications to liver segmentation in CT scans. Comput. Math. Methods Med. 2014, 2014, 182909.
- Mostafa, A.; Fouad, A.; Abd Elfattah, M.; Hassanien, A.E.; Hefny, H.; Zhu, S.Y.; Schaefer, G. CT liver segmentation using artificial bee colony optimisation. Procedia Comput. Sci. 2015, 60, 1622–1630.
- Dou, Q.; Chen, H.; Jin, Y.; Yu, L.; Qin, J.; Heng, P.A. 3D deeply supervised network for automatic liver segmentation from CT volumes. In Proceedings of the 19th International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Athens, Greece, 17–21 October 2016; Springer: Berlin/Heidelberg, Germany, 2016; pp. 149–157.
- Hiraman, A. Liver Segmentation Using 3D CT Scans. Ph.D. Thesis, University of Kwazulu-Natal, Durban, South Africa, 2018.
- Wang, K.; Mamidipalli, A.; Retson, T.; Bahrami, N.; Hasenstab, K.; Blansit, K.; Bass, E.; Delgado, T.; Cunha, G.; Middleton, M.S.; et al. Automated CT and MRI liver segmentation and biometry using a generalized convolutional neural network. Radiol. Artif. Intell. 2019, 1, 180022.
- Almotairi, S.; Kareem, G.; Aouf, M.; Almutairi, B.; Salem, M.A.M. Liver tumor segmentation in CT scans using modified SegNet. Sensors 2020, 20, 1516.
- Soler, L.; Hostettler, A.; Agnus, V.; Charnoz, A.; Fasquel, J.; Moreau, J.; Osswald, A.; Bouhadjar, M.; Marescaux, J. 3D Image Reconstruction for Comparison of Algorithm Database: A Patient Specific Anatomical and Medical Image Database; Technical Report; IRCAD: Strasbourg, France, 2010.
- Ayalew, Y.A.; Fante, K.A.; Mohammed, M.A. Modified U-Net for liver cancer segmentation from computed tomography images with a new class balancing method. BMC Biomed. Eng. 2021, 3, 4.
- LiTS Challenge Dataset. Available online: https://competitions.codalab.org/competitions/17094 (accessed on 27 November 2023).
- Scicluna, D. Automatic Segmentation of Healthy Liver in Abdominal Computed Tomography Scans. Master’s Thesis, University of Malta, Msida, Malta, 2022.
- Kavur, A.E.; Gezer, N.S.; Barış, M.; Aslan, S.; Conze, P.H.; Groza, V.; Pham, D.D.; Chatterjee, S.; Ernst, P.; Özkan, S.; et al. CHAOS Challenge-combined (CT-MR) healthy abdominal organ segmentation. Med. Image Anal. 2021, 69, 101950.
- Ezzat, K.A.; Omran, L.N.; El Seddawy, A.I.B. Automatic liver segmentation in computed tomography scans using deep semantic segmentation. Bull. Electr. Eng. Inform. 2023, 12, 250–256.
- Shao, J.; Luan, S.; Ding, Y.; Xue, X.; Zhu, B.; Wei, W. Attention Connect Network for Liver Tumor Segmentation from CT and MRI Images. Technol. Cancer Res. Treat. 2024, 23, 15330338231219366.
- Maurício, J.; Domingues, I.; Bernardino, J. Comparing Vision Transformers and Convolutional Neural Networks for Image Classification: A Literature Review. Appl. Sci. 2023, 13, 5521.
- Al-Saeed, Y.; Gab-Allah, W.; Elmogy, M. Hepatic tumors diagnosis system based on fuzzy c-means using computed tomography images. Res. Sq. Prepr. 2022.
- MICCAI-Sliver07. Available online: https://sliver07.grand-challenge.org/ (accessed on 27 November 2023).
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25.
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv 2015, arXiv:1505.04597.
This entry is offline, you can click here to edit this entry!
