The role of capsule endoscopy and enteroscopy in managing various small-bowel pathologies is well-established. However, their broader application has been hampered mainly by their lengthy reading times. As a result, there is a growing interest in employing artificial intelligence (AI) in these diagnostic and therapeutic procedures, driven by the prospect of overcoming some major limitations and enhancing healthcare efficiency, while maintaining high accuracy levels.
- artificial intelligence
- convolutional neural network
- deep learning
- small bowel
- capsule endoscopy
1. Introduction
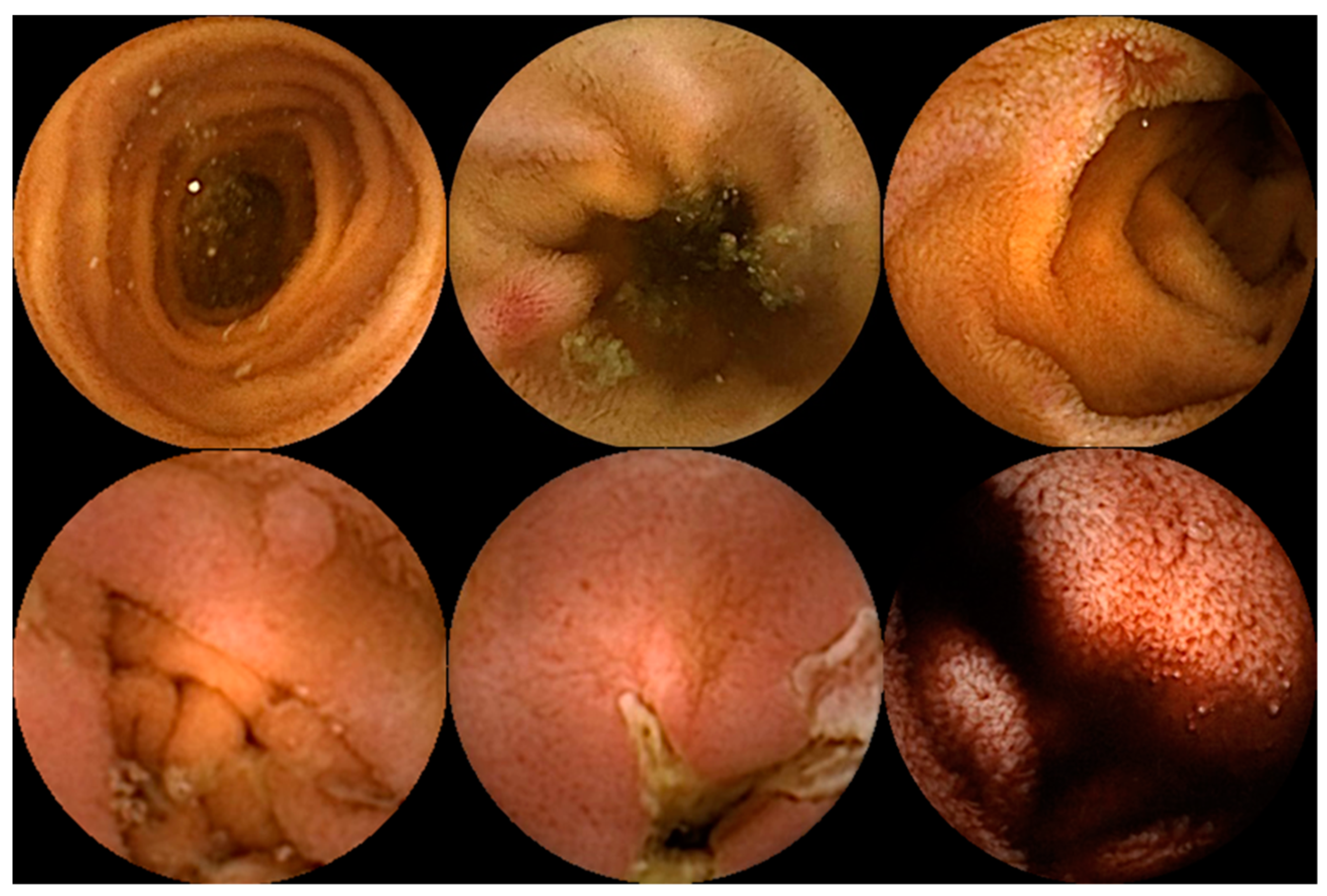
Capsule endoscopy allows for a non-invasive and painless evaluation of small bowel (SB) mucosa, essentially being a diagnostic modality [1][2]. This exam is fundamental to the diagnosis of obscure gastrointestinal bleeding (OGIB) but also the study of Crohn’s disease (CD), SB tumors, celiac disease (CeD) (extent and severity), and others [1][2][3], as illustrated in Figure 1. However, it is essential to note that CE has some drawbacks. Among these is the dependence on the examiner’s clinical experience and the time and labor involved in the image review process (previous series have reported reading times of over 40 to 50 min), which makes it a task prone to error [4][5]. Therefore, artificial intelligence (AI) will probably contribute to minimizing these limitations and increase its potential. Nowadays, this topic is becoming more popular, resulting in an increasing number of recent research articles dedicated to it.
2. Artificial Intelligence and Obscure Gastrointestinal Bleeding
3. Artificial Intelligence and Vascular Lesions
4. Artificial Intelligence and Protruding Lesions
5. Artificial Intelligence and Pleomorphic Lesion Detection
6. Artificial Intelligence and Small-Bowel Compartmentalization
7. Artificial Intelligence and Celiac Disease
8. Artificial Intelligence and Inflammatory Bowel Activity
9. Artificial Intelligence and Small-Bowel Cleansing
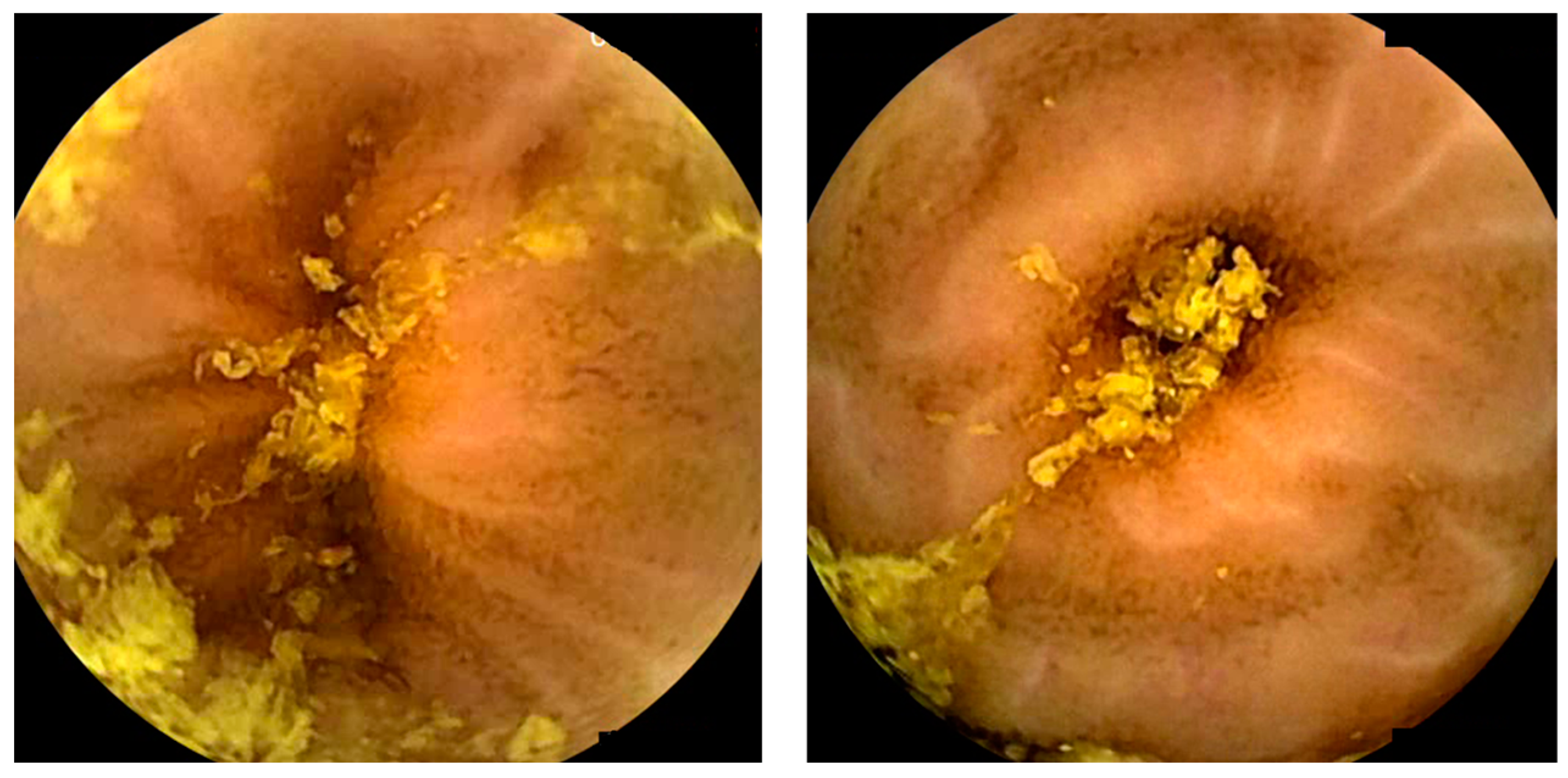
10. Miscellaneous—Artificial Intelligence and Hookworms/Functional Bowel disorders
This entry is adapted from the peer-reviewed paper 10.3390/diagnostics14030291
References
- Okagawa, Y.; Abe, S.; Yamada, M.; Oda, I.; Saito, Y. Artificial Intelligence in Endoscopy. Dig. Dis. Sci. 2022, 67, 1553–1572.
- Cortegoso Valdivia, P.; Skonieczna-Zydecka, K.; Elosua, A.; Sciberras, M.; Piccirelli, S.; Rullan, M.; Tabone, T.; Gawel, K.; Stachowski, A.; Leminski, A.; et al. Indications, Detection, Completion and Retention Rates of Capsule Endoscopy in Two Decades of Use: A Systematic Review and Meta-Analysis. Diagnostics 2022, 12, 1105.
- Ciaccio, E.J.; Tennyson, C.A.; Bhagat, G.; Lewis, S.K.; Green, P.H. Classification of videocapsule endoscopy image patterns: Comparative analysis between patients with celiac disease and normal individuals. Biomed. Eng. Online 2010, 9, 44.
- Majtner, T.; Brodersen, J.B.; Herp, J.; Kjeldsen, J.; Halling, M.L.; Jensen, M.D. A deep learning framework for autonomous detection and classification of Crohn’s disease lesions in the small bowel and colon with capsule endoscopy. Endosc. Int. Open 2021, 9, E1361–E1370.
- Mascarenhas, M.; Cardoso, H.; Macedo, G. Artificial Intelligence in Capsule Endoscopy: A Gamechanger for a Groundbreaking Technique; Elsevier: Amsterdam, The Netherlands, 2023.
- Awadie, H.; Zoabi, A.; Gralnek, I.M. Obscure-overt gastrointestinal bleeding: A review. Pol. Arch. Intern. Med. 2022, 132, 16253.
- Patel, A.; Vedantam, D.; Poman, D.S.; Motwani, L.; Asif, N. Obscure Gastrointestinal Bleeding and Capsule Endoscopy: A Win-Win Situation or Not? Cureus 2022, 14, e27137.
- Jackson, C.S.; Strong, R. Gastrointestinal Angiodysplasia: Diagnosis and Management. Gastrointest. Endosc. Clin. N. Am. 2017, 27, 51–62.
- Afonso, J.; Saraiva, M.M.; Ferreira, J.P.S.; Ribeiro, T.; Cardoso, H.; Macedo, G. Performance of a convolutional neural network for automatic detection of blood and hematic residues in small bowel lumen. Dig. Liver Dis. 2021, 53, 654–657.
- Pan, G.; Yan, G.; Qiu, X.; Cui, J. Bleeding detection in Wireless Capsule Endoscopy based on Probabilistic Neural Network. J. Med. Syst. 2011, 35, 1477–1484.
- Fu, Y.; Zhang, W.; Mandal, M.; Meng, M.Q. Computer-aided bleeding detection in WCE video. IEEE J. Biomed. Health Inf. 2014, 18, 636–642.
- Xiao, J.; Meng, M.Q. A deep convolutional neural network for bleeding detection in Wireless Capsule Endoscopy images. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2016, 2016, 639–642.
- Fan, S.; Xu, L.; Fan, Y.; Wei, K.; Li, L. Computer-aided detection of small intestinal ulcer and erosion in wireless capsule endoscopy images. Phys. Med. Biol. 2018, 63, 165001.
- Aoki, T.; Yamada, A.; Aoyama, K.; Saito, H.; Tsuboi, A.; Nakada, A.; Niikura, R.; Fujishiro, M.; Oka, S.; Ishihara, S.; et al. Automatic detection of erosions and ulcerations in wireless capsule endoscopy images based on a deep convolutional neural network. Gastrointest. Endosc. 2019, 89, 357–363.e352.
- Wang, S.; Xing, Y.; Zhang, L.; Gao, H.; Zhang, H. A systematic evaluation and optimization of automatic detection of ulcers in wireless capsule endoscopy on a large dataset using deep convolutional neural networks. Phys. Med. Biol. 2019, 64, 235014.
- Aoki, T.; Yamada, A.; Aoyama, K.; Saito, H.; Fujisawa, G.; Odawara, N.; Kondo, R.; Tsuboi, A.; Ishibashi, R.; Nakada, A.; et al. Clinical usefulness of a deep learning-based system as the first screening on small-bowel capsule endoscopy reading. Dig. Endosc. 2020, 32, 585–591.
- Aoki, T.; Yamada, A.; Kato, Y.; Saito, H.; Tsuboi, A.; Nakada, A.; Niikura, R.; Fujishiro, M.; Oka, S.; Ishihara, S.; et al. Automatic detection of blood content in capsule endoscopy images based on a deep convolutional neural network. J. Gastroenterol. Hepatol. 2020, 35, 1196–1200.
- Ghosh, T.; Chakareski, J. Deep Transfer Learning for Automated Intestinal Bleeding Detection in Capsule Endoscopy Imaging. J. Digit. Imaging 2021, 34, 404–417.
- Mascarenhas Saraiva, M.J.; Afonso, J.; Ribeiro, T.; Ferreira, J.; Cardoso, H.; Andrade, A.P.; Parente, M.; Natal, R.; Mascarenhas Saraiva, M.; Macedo, G. Deep learning and capsule endoscopy: Automatic identification and differentiation of small bowel lesions with distinct haemorrhagic potential using a convolutional neural network. BMJ Open Gastroenterol. 2021, 8.
- Vieira, P.M.; Silva, C.P.; Costa, D.; Vaz, I.F.; Rolanda, C.; Lima, C.S. Automatic Segmentation and Detection of Small Bowel Angioectasias in WCE Images. Ann. Biomed. Eng. 2019, 47, 1446–1462.
- Vieira, P.M.; Goncalves, B.; Goncalves, C.R.; Lima, C.S. Segmentation of angiodysplasia lesions in WCE images using a MAP approach with Markov Random Fields. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2016, 2016, 1184–1187.
- Noya, F.; Alvarez-Gonzalez, M.A.; Benitez, R. Automated angiodysplasia detection from wireless capsule endoscopy. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2017, 2017, 3158–3161.
- Leenhardt, R.; Vasseur, P.; Li, C.; Saurin, J.C.; Rahmi, G.; Cholet, F.; Becq, A.; Marteau, P.; Histace, A.; Dray, X. A neural network algorithm for detection of GI angiectasia during small-bowel capsule endoscopy. Gastrointest. Endosc. 2019, 89, 189–194.
- Tsuboi, A.; Oka, S.; Aoyama, K.; Saito, H.; Aoki, T.; Yamada, A.; Matsuda, T.; Fujishiro, M.; Ishihara, S.; Nakahori, M.; et al. Artificial intelligence using a convolutional neural network for automatic detection of small-bowel angioectasia in capsule endoscopy images. Dig. Endosc. 2020, 32, 382–390.
- Chu, Y.; Huang, F.; Gao, M.; Zou, D.W.; Zhong, J.; Wu, W.; Wang, Q.; Shen, X.N.; Gong, T.T.; Li, Y.Y.; et al. Convolutional neural network-based segmentation network applied to image recognition of angiodysplasias lesion under capsule endoscopy. World J. Gastroenterol. 2023, 29, 879–889.
- Van de Bruaene, C.; De Looze, D.; Hindryckx, P. Small bowel capsule endoscopy: Where are we after almost 15 years of use? World J. Gastrointest. Endosc. 2015, 7, 13–36.
- Mascarenhas Saraiva, M.; Afonso, J.; Ribeiro, T.; Ferreira, J.; Cardoso, H.; Andrade, P.; Gonçalves, R.; Cardoso, P.; Parente, M.; Jorge, R.; et al. Artificial intelligence and capsule endoscopy: Automatic detection of enteric protruding lesions using a convolutional neural network. Rev. Esp. Enferm. Dig. 2023, 115, 75–79.
- Barbosa, D.J.; Ramos, J.; Lima, C.S. Detection of small bowel tumors in capsule endoscopy frames using texture analysis based on the discrete wavelet transform. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2008, 2008, 3012–3015.
- Barbosa, D.C.; Roupar, D.B.; Ramos, J.C.; Tavares, A.C.; Lima, C.S. Automatic small bowel tumor diagnosis by using multi-scale wavelet-based analysis in wireless capsule endoscopy images. Biomed. Eng. Online 2012, 11, 3.
- Li, B.; Meng, M.Q.; Xu, L. A comparative study of shape features for polyp detection in wireless capsule endoscopy images. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2009, 2009, 3731–3734.
- Li, B.P.; Meng, M.Q. Comparison of several texture features for tumor detection in CE images. J. Med. Syst. 2012, 36, 2463–2469.
- Li, B.; Meng, M.Q. Tumor recognition in wireless capsule endoscopy images using textural features and SVM-based feature selection. IEEE Trans. Inf. Technol. Biomed. 2012, 16, 323–329.
- Vieira, P.M.; Freitas, N.R.; Valente, J.; Vaz, I.F.; Rolanda, C.; Lima, C.S. Automatic detection of small bowel tumors in wireless capsule endoscopy images using ensemble learning. Med. Phys. 2020, 47, 52–63.
- Vieira, P.M.; Ramos, J.; Lima, C.S. Automatic detection of small bowel tumors in endoscopic capsule images by ROI selection based on discarded lightness information. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2015, 2015, 3025–3028.
- Yuan, Y.; Meng, M.Q. Deep learning for polyp recognition in wireless capsule endoscopy images. Med. Phys. 2017, 44, 1379–1389.
- Saito, H.; Aoki, T.; Aoyama, K.; Kato, Y.; Tsuboi, A.; Yamada, A.; Fujishiro, M.; Oka, S.; Ishihara, S.; Matsuda, T.; et al. Automatic detection and classification of protruding lesions in wireless capsule endoscopy images based on a deep convolutional neural network. Gastrointest. Endosc. 2020, 92, 144–151.e141.
- Hwang, Y.; Lee, H.H.; Park, C.; Tama, B.A.; Kim, J.S.; Cheung, D.Y.; Chung, W.C.; Cho, Y.S.; Lee, K.M.; Choi, M.G.; et al. Improved classification and localization approach to small bowel capsule endoscopy using convolutional neural network. Dig. Endosc. 2021, 33, 598–607.
- Ding, Z.; Shi, H.; Zhang, H.; Meng, L.; Fan, M.; Han, C.; Zhang, K.; Ming, F.; Xie, X.; Liu, H.; et al. Gastroenterologist-Level Identification of Small-Bowel Diseases and Normal Variants by Capsule Endoscopy Using a Deep-Learning Model. Gastroenterology 2019, 157, 1044–1054.e1045.
- Otani, K.; Nakada, A.; Kurose, Y.; Niikura, R.; Yamada, A.; Aoki, T.; Nakanishi, H.; Doyama, H.; Hasatani, K.; Sumiyoshi, T.; et al. Automatic detection of different types of small-bowel lesions on capsule endoscopy images using a newly developed deep convolutional neural network. Endoscopy 2020, 52, 786–791.
- Aoki, T.; Yamada, A.; Kato, Y.; Saito, H.; Tsuboi, A.; Nakada, A.; Niikura, R.; Fujishiro, M.; Oka, S.; Ishihara, S.; et al. Automatic detection of various abnormalities in capsule endoscopy videos by a deep learning-based system: A multicenter study. Gastrointest. Endosc. 2021, 93, 165–173.e161.
- Vieira, P.M.; Freitas, N.R.; Lima, V.B.; Costa, D.; Rolanda, C.; Lima, C.S. Multi-pathology detection and lesion localization in WCE videos by using the instance segmentation approach. Artif. Intell. Med. 2021, 119, 102141.
- Wang, C.; Luo, Z.; Liu, X.; Bai, J.; Liao, G. Organic Boundary Location Based on Color-Texture of Visual Perception in Wireless Capsule Endoscopy Video. J. Healthc. Eng. 2018, 2018, 3090341.
- Raiteri, A.; Granito, A.; Giamperoli, A.; Catenaro, T.; Negrini, G.; Tovoli, F. Current guidelines for the management of celiac disease: A systematic review with comparative analysis. World J. Gastroenterol. 2022, 28, 154–175.
- Zhou, T.; Han, G.; Li, B.N.; Lin, Z.; Ciaccio, E.J.; Green, P.H.; Qin, J. Quantitative analysis of patients with celiac disease by video capsule endoscopy: A deep learning method. Comput. Biol. Med. 2017, 85, 1–6.
- Koh, J.E.W.; Hagiwara, Y.; Oh, S.L.; Tan, J.H.; Ciaccio, E.J.; Green, P.H.; Lewis, S.K.; Acharya, U.R. Automated diagnosis of celiac disease using DWT and nonlinear features with video capsule endoscopy images. Future Gener. Comput. Syst. 2018, 90, 86–93.
- Wang, X.; Qian, H.; Ciaccio, E.J.; Lewis, S.K.; Bhagat, G.; Green, P.H.; Xu, S.; Huang, L.; Gao, R.; Liu, Y. Celiac disease diagnosis from videocapsule endoscopy images with residual learning and deep feature extraction. Comput. Methods Programs Biomed. 2020, 187, 105236.
- Stoleru, C.A.; Dulf, E.H.; Ciobanu, L. Automated detection of celiac disease using Machine Learning Algorithms. Sci. Rep. 2022, 12, 4071.
- Chetcuti Zammit, S.; McAlindon, M.E.; Greenblatt, E.; Maker, M.; Siegelman, J.; Leffler, D.A.; Yardibi, O.; Raunig, D.; Brown, T.; Sidhu, R. Quantification of Celiac Disease Severity Using Video Capsule Endoscopy: A Comparison of Human Experts and Machine Learning Algorithms. Curr. Med. Imaging 2023, 19, 1455–1662.
- Goran, L.; Negreanu, A.M.; Stemate, A.; Negreanu, L. Capsule endoscopy: Current status and role in Crohn’s disease. World J. Gastrointest. Endosc. 2018, 10, 184–192.
- Lamb, C.A.; Kennedy, N.A.; Raine, T.; Hendy, P.A.; Smith, P.J.; Limdi, J.K.; Hayee, B.; Lomer, M.C.E.; Parkes, G.C.; Selinger, C.; et al. British Society of Gastroenterology consensus guidelines on the management of inflammatory bowel disease in adults. Gut 2019, 68, s1–s106.
- Klang, E.; Barash, Y.; Margalit, R.Y.; Soffer, S.; Shimon, O.; Albshesh, A.; Ben-Horin, S.; Amitai, M.M.; Eliakim, R.; Kopylov, U. Deep learning algorithms for automated detection of Crohn’s disease ulcers by video capsule endoscopy. Gastrointest. Endosc. 2020, 91, 606–613.e602.
- Takenaka, K.; Kawamoto, A.; Okamoto, R.; Watanabe, M.; Ohtsuka, K. Artificial intelligence for endoscopy in inflammatory bowel disease. Intest. Res. 2022, 20, 165–170.
- Barash, Y.; Azaria, L.; Soffer, S.; Margalit Yehuda, R.; Shlomi, O.; Ben-Horin, S.; Eliakim, R.; Klang, E.; Kopylov, U. Ulcer severity grading in video capsule images of patients with Crohn’s disease: An ordinal neural network solution. Gastrointest. Endosc. 2021, 93, 187–192.
- Klang, E.; Grinman, A.; Soffer, S.; Margalit Yehuda, R.; Barzilay, O.; Amitai, M.M.; Konen, E.; Ben-Horin, S.; Eliakim, R.; Barash, Y.; et al. Automated Detection of Crohn’s Disease Intestinal Strictures on Capsule Endoscopy Images Using Deep Neural Networks. J. Crohns Colitis 2021, 15, 749–756.
- Noorda, R.; Nevárez, A.; Colomer, A.; Pons Beltrán, V.; Naranjo, V. Automatic evaluation of degree of cleanliness in capsule endoscopy based on a novel CNN architecture. Sci. Rep. 2020, 10, 17706.
- Ju, J.; Oh, H.S.; Lee, Y.J.; Jung, H.; Lee, J.H.; Kang, B.; Choi, S.; Kim, J.H.; Kim, K.O.; Chung, Y.J. Clean mucosal area detection of gastroenterologists versus artificial intelligence in small bowel capsule endoscopy. Medicine 2023, 102, e32883.
- Rosa, B.; Margalit-Yehuda, R.; Gatt, K.; Sciberras, M.; Girelli, C.; Saurin, J.C.; Valdivia, P.C.; Cotter, J.; Eliakim, R.; Caprioli, F.; et al. Scoring systems in clinical small-bowel capsule endoscopy: All you need to know! Endosc. Int. Open 2021, 9, E802–E823.
- Van Weyenberg, S.J.; De Leest, H.T.; Mulder, C.J. Description of a novel grading system to assess the quality of bowel preparation in video capsule endoscopy. Endoscopy 2011, 43, 406–411.
- Ponte, A.; Pinho, R.; Rodrigues, A.; Silva, J.; Rodrigues, J.; Carvalho, J. Validation of the computed assessment of cleansing score with the Mirocam® system. Rev. Esp. Enferm. Dig. 2016, 108, 709–715.
- Abou Ali, E.; Histace, A.; Camus, M.; Gerometta, R.; Becq, A.; Pietri, O.; Nion-Larmurier, I.; Li, C.; Chaput, U.; Marteau, P.; et al. Development and validation of a computed assessment of cleansing score for evaluation of quality of small-bowel visualization in capsule endoscopy. Endosc. Int. Open 2018, 6, E646–E651.
- Oumrani, S.; Histace, A.; Abou Ali, E.; Pietri, O.; Becq, A.; Houist, G.; Nion-Larmurier, I.; Camus, M.; Florent, C.; Dray, X. Multi-criterion, automated, high-performance, rapid tool for assessing mucosal visualization quality of still images in small bowel capsule endoscopy. Endosc. Int. Open 2019, 7, E944–E948.
- Leenhardt, R.; Souchaud, M.; Houist, G.; Le Mouel, J.P.; Saurin, J.C.; Cholet, F.; Rahmi, G.; Leandri, C.; Histace, A.; Dray, X. A neural network-based algorithm for assessing the cleanliness of small bowel during capsule endoscopy. Endoscopy 2021, 53, 932–936.
- Nam, J.H.; Hwang, Y.; Oh, D.J.; Park, J.; Kim, K.B.; Jung, M.K.; Lim, Y.J. Development of a deep learning-based software for calculating cleansing score in small bowel capsule endoscopy. Sci. Rep. 2021, 11, 4417.
- Ju, J.W.; Jung, H.; Lee, Y.J.; Mun, S.W.; Lee, J.H. Semantic Segmentation Dataset for AI-Based Quantification of Clean Mucosa in Capsule Endoscopy. Medicina 2022, 58, 397.
- Ribeiro, T.; Mascarenhas Saraiva, M.J.; Afonso, J.; Cardoso, P.; Mendes, F.; Martins, M.; Andrade, A.P.; Cardoso, H.; Mascarenhas Saraiva, M.; Ferreira, J.; et al. Design of a Convolutional Neural Network as a Deep Learning Tool for the Automatic Classification of Small-Bowel Cleansing in Capsule Endoscopy. Medicina 2023, 59, 810.
- Houdeville, C.; Leenhardt, R.; Souchaud, M.; Velut, G.; Carbonell, N.; Nion-Larmurier, I.; Nuzzo, A.; Histace, A.; Marteau, P.; Dray, X. Evaluation by a Machine Learning System of Two Preparations for Small Bowel Capsule Endoscopy: The BUBS (Burst Unpleasant Bubbles with Simethicone) Study. J. Clin. Med. 2022, 11, 2822.
- Wu, X.; Chen, H.; Gan, T.; Chen, J.; Ngo, C.W.; Peng, Q. Automatic Hookworm Detection in Wireless Capsule Endoscopy Images. IEEE Trans. Med. Imaging 2016, 35, 1741–1752.
- Gan, T.; Yang, Y.; Liu, S.; Zeng, B.; Yang, J.; Deng, K.; Wu, J.; Yang, L. Automatic Detection of Small Intestinal Hookworms in Capsule Endoscopy Images Based on a Convolutional Neural Network. Gastroenterol. Res. Pr. 2021, 2021, 5682288.
- Spyridonos, P.; Vilariño, F.; Vitrià, J.; Azpiroz, F.; Radeva, P. Anisotropic feature extraction from endoluminal images for detection of intestinal contractions. Med. Image Comput. Comput. Assist. Interv. 2006, 9, 161–168.
- Malagelada, C.; De Iorio, F.; Azpiroz, F.; Accarino, A.; Segui, S.; Radeva, P.; Malagelada, J.R. New insight into intestinal motor function via noninvasive endoluminal image analysis. Gastroenterology 2008, 135, 1155–1162.
