Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Blastocystis is the most common gastrointestinal protist found in humans and animals. Although the clinical significance of Blastocystis remains unclear, the organism is increasingly being viewed as a commensal member of the gut microbiome.
- Blastocystis
- gut microbiome
- microbiome modulation
1. Introduction
Blastocystis is one of the most common microbial eukaryotes in the gastrointestinal tracts of humans and animals. Based on small subunit ribosomal RNA (SSUrRNA), the genus is composed of many genetically distinct subtypes (STs) that most likely represent separate species. The current taxonomy of Blastocystis is as follows: the kingdom Sar, the phylum Stramenopiles, the class Bigyra, the order Opalinata, the family Blastocystidae and the genus Blastocystis; species are not applicable [1]. Stramenopiles comprise over 100,000 species distributed across 21 classes. The majority of described species are diatoms, followed by brown algae, chrysophytes, xanthophytes, and oomycetes. However, unlike most other members of Stramenopiles, Blastocystis is neither flagellated nor motile [2].
Blastocystis has a global distribution; however, higher frequencies have been reported in developing countries because of poor hygiene, animal handling, or the fecal contamination of food and water [3][4]. The range of genetic diversity in Blastocystis is considerably high, and recently, at least 42 STs were identified from various hosts, relying on small subunit ribosomal RNA gene (SSU rRNA) polymorphisms [5][6][7]. In fact, one of the most significant current discussions is the number of STs and the identification of novel subtypes [8]. The genome of Blastocystis ST7 was the first to be sequenced in 2011, with data from ST1, ST2, ST4, ST6, ST8, and ST9 becoming available later at various stages of annotation [9][10]. Despite sharing common core genes, some important features, including genome sizes, intron numbers, guanine–cytosine (GC) contents, and gene contents, vary among subtypes [11].
The role of Blastocystis in the development of gastrointestinal diseases has also been much disputed despite a considerable number of studies [12][13][14]. Blastocystis infection has been associated with non-specific gastrointestinal symptoms such as abdominal pain, diarrhea, nausea, vomiting, bloating, and anorexia, as well as less frequent dermatological complaints like urticaria and severe itching [2][15][16][17]. In vitro studies on Blastocystis pathogenesis demonstrated that it can attach to the intestinal mucosa, increase intestinal permeability by secreting cysteine proteases, degrade secretory immunoglobulin A (IgA), induce the secretion of inflammatory cytokines such as interleukin-8, and cause the apoptosis of host cells [1][18]. In general, the prevalence of Blastocystis has been reported to be higher in healthy populations compared to individuals with ulcerative colitis (UC) or irritable bowel syndrome (IBS) [19][20]. Blastocystis resides in the human intestine for a long period of time without causing any symptoms, encouraging the question of whether it should be considered a pathogen or a commensal microorganism [21][22]. Nonetheless, the eradication of Blastocystis is considered necessary in cases where it is the sole protist agent and the patient’s complaints persist [23].
The gut microbiome refers to the collection of bacteria, viruses, archaea, and eukaryotes that colonize the gastrointestinal tract, primarily the large intestine. This highly dynamic and complex ecosystem plays a crucial role in maintaining human health and has various physiological functions. It is currently accepted that the human gut microbiome is first acquired and established before or during birth, with the mode of delivery, ethnicity, and host genetics playing roles in its composition [24][25]. In addition, various external factors such as diet, nutritional status, prenatal events, geographical location, antibiotic treatment, and age contribute to establishing the gut microbiome throughout human life [26][27][28][29][30]. The microbiome reaches a “balanced” state with high taxonomic microbial diversity and richness in the following years of life, forming a commensal relationship with the host [31]. The Human Microbiome Project (HMP) and the Metagenomics of the Human Intestinal Tract (MetaHIT) project, as well as the development of novel technologies such as 16S rRNA gene metabarcoding, have improved our understanding [32][33]. The study of the gut microbiome has become a major area of interest in various disciplines. These days, some define the microbiome as a novel multicellular “organ” which interacts closely with its host [34]. The gut microbiome has numerous important functions including digestion, nutrient production, immune system regulation, gut barrier function for pathogens, and the regulation of metabolic activities; therefore, maintaining a healthy and diverse gut microbiome is essential for overall well-being [35][36]. The term “dysbiosis” can be defined as a persistent imbalance in the gut microbial community and can lead to various chronic conditions. Integrative analyses of the gut microbiome in humans and laboratory animals have offered possible relationships with many chronic diseases such as autoimmune disorders, obesity, diabetes, IBS, metabolic syndrome, depression, and allergy [37][38][39][40][41][42].
Single-celled eukaryotes constitute an important and heterogeneous group within the human intestinal microbiota. A major discussion point revolves around the categorization of these species as pathogenic, commensal, beneficial, or opportunistic pathogens. The well-known gut-related protozoa in humans are Blastocystis, Dientamoeba fragilis, Giardia intestinalis, Entamoeba histolytica, and Cryptosporidium spp. Among these, the last three significantly contribute to acute gastroenteritis and diarrheal diseases on a global scale [43]. However, many intestinal protist species, such as Endolimax nana, Entamoeba polecki, Iodamoeba butschlii, and Chilomastix mesnili, are non-pathogenic and might even be beneficial inhabitants of the gut [44].
2. Blastocystis and the Gut Microbiome
2.1. The Effect of Blastocystis on Gut Microbiome Modulation
Blastocystis colonization is thought to be related to changes in the gut bacterial microbiome [45]. Recent studies indicate that Blastocystis infection may be associated with alterations in the abundances of both beneficial and harmful intestinal bacteria. Research on the relationship between asymptomatic Blastocystis infection and intestinal bacterial composition is ongoing, although this association still needs to be fully understood [46][47][48]. Behboud et al. have reported that the mean relative abundances of Bifidobacterium and Lactobacillus/Enterococcus (beneficial bacteria) groups and Peptostreptococcus productus and Escherichia coli (harmful bacteria) were upregulated significantly, while the relative abundances of Bacteroides fragilis (B. fragilis) and Enterococcus sp. were downregulated considerably in those with Blastocystis compared to a control group [47].
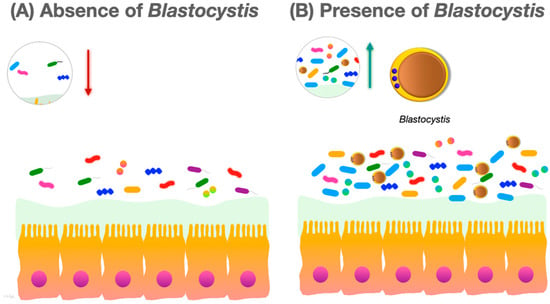
Many studies report that colonization with Blastocystis is associated with increased diversity of the human intestinal bacterial microbiota (Figure 1).

Figure 1. (A) In the absence of Blastocystis, the gut microbiota species richness and community evenness are lower; (B) in the presence of Blastocystis, the species richness and community evenness of the gut microbiota increases.
There are limited data in the literature on the relationships between Blastocystis STs and the gut microbiota. Blastocystis ST1 is one of the most commonly found STs in humans worldwide [49][50][51]. Some studies have demonstrated that ST1 has beneficial effects on the host gut microbiome and immune system. Deng et al. showed that colonization with Blastocystis ST1 could increase the levels of Alloprevotella and Akkermansia, which are beneficial bacteria for gut health, in a murine model [52]. Blastocystis ST3, another very common subtype, has been shown to cause an increase in beneficial bacteria such as Bacillota (syn. Firmicutes) and Bacteroidota (syn. Bacteroidetes) in the host gut microbiome, and it has been reported that this may indirectly be beneficial to the host immune response [53].
It is considered that the composition of the gut microbiome plays a crucial role in the pathogenesis of certain diseases such as IBD, which is a chronic inflammation of the gastrointestinal tract. Although some studies have reported a high prevalence of Blastocystis in patients with IBD, the relationship between the organism and the disease is still controversial [54].
2.2. The Effect of Blastocystis on Immune Modulation
The gut microbiome plays an essential role in the health and disease status of the host. It is now known that it contributes significantly to the pathogenesis of autoimmune diseases, with the deterioration of the gut microbiome being linked to the dysregulation of the immune system [55]. The pathogenic potential of Blastocystis, its clinical significance, and its potential effects on the host immune system are still debated [46]. Whether Blastocystis is pathogenic or non-pathogenic depends on factors such as its interaction with the human gut microbiome, the subtype, and the human immune response regulators or modulators involved [56]. While Blastocystis colonizes the human gut and does not cause any infection, this situation can change in the event of a disruption to the immune system or gut microbiome balance [57]. An investigation of the metabolic profiles of Blastocystis carriers and non-carriers revealed differential levels of certain amino acids (Ala, Gly, His, Ile, Met, Thr, Try, and Val) in fecal samples collected from individuals from different countries. These amino acids are considered inflammatory biomarkers if their abundance in the gut is increased. The decrease in these amino acids in Blastocystis-positive individuals may indicate that the organism assumes an anti-inflammatory role in the intestine [58].
One of the areas of interest in Blastocystis research, along with recent work, is the insight into its potential to modulate the host immune system. Research suggests that Blastocystis infection can elicit various immune responses, including both pro-inflammatory and anti-inflammatory responses. However, the exact nature and significance of these immune responses are not fully understood [59].
The gut microbial ecosystem is crucial for the modulation and regulation of the immune system [60][61]. Mucin, a thick and sticky glycoprotein, is produced by goblet cells that secrete mucus in the body, especially in the gastrointestinal tract. Cysteine proteases, produced by pathogenic parasites, cause the breakdown of mucin, creating gaps between colon epithelial cells and thus making the invasion of the underlying host tissue possible [62]. The gut microbiome can support the development of T regulatory cells (Tregs) by producing short-chain fatty acids (SCFAs) and regulating Th2 immune responses during parasite infection. More specifically, Blastocystis ST4 has been associated with increased abundances of bacteria such as Akkermansia spp. and SCFA-producing bacteria associated with increased SCFA production, which can provide energy to goblet cells [44][63]. These data indicate subtype-specific effects of Blastocystis on immune modulation.
Immunoglobulin A is crucial in the mucosal defense of the gastrointestinal tract as it provides immune protection against microbial pathogens [64]. The release of cysteine protease by Blastocystis ST7 and aspartic proteases by ST4 has been shown to mediate the degradation of IgA and subsequently modulate the host immune response [57][65]. It has been demonstrated that Blastocystis ST4 cysteine proteases induce the upstream synthesis of interleukin (IL)-8 through the nuclear factor-κB (NF-κB) pathway [66]. An increase in the proinflammatory chemokine IL-8 and granulocyte–macrophage colony-stimulating factor (GM-CSF) in human colon carcinoma cells with a Blastocystis ST1 co-culture has been reported [2].
2.3. The Interaction of Blastocystis and the Gut Microbiome in Autoimmune Diseases
Autoimmune diseases (ADs) occur when cells of the immune system attack the host’s cells and tissues, resulting in chronic inflammation. In the last decade, it has become known that environmental factors trigger ADs in genetically predisposed individuals [67]. The gut microbiome, which consists of trillions of microorganisms inhabiting the gastrointestinal tract, plays a critical role in regulating the immune system and maintaining gut health. It has been suggested that imbalances, either an increase or decrease in the specific taxa of the gut microbiome, may contribute to the development of ADs [68]. A disturbed balance in the gut microbiome may be associated not only with intestinal ADs (IBD, IBS, celiac disease, and autoimmune gastritis, etc.) but also with extra-intestinal ADs (multiple sclerosis, rheumatoid arthritis (RA), type 1 diabetes, and systemic lupus erythematosus (SLE)).
Spondyloarthritis comprises a group of rheumatic diseases with differential clinical features, such as ankylosing spondylitis (AS), reactive arthritis (ReA), and psoriatic arthritis (PsA), along with inflammatory bowel disease-associated SpA, uveitis, and dermatological and gastroenterological involvement [69]. A prevalent feature in many inflammatory diseases, including SpA, is gut microbial dysbiosis. Patients with SpA showed a decreased fecal abundance of Faecalibacterium prausnitzii and an increase in B. fragilis [70]. Regarding the gut microbiome composition of SpA patients, the main results from a meta-analysis showed increased frequencies of Bacteroidaceae and Enterobacteriaceae in the phylum Pseudomonadota (syn Proteobacteria), while the gut microbiome diversity in the phylum Bacteroidota (syn Bacteroidetes) showed decreases in Bacteroidales and Akkermansia [71].
Irritable bowel syndrome is a common functional gastrointestinal disorder characterized by abdominal pain, discomfort during defecation, and changes in the gut microbiome [72]. Some studies have reported that the gut microbiome of IBS patients had a significantly increased number of bacteria in the families Enterobacteriaceae and Bacteroides compared to healthy controls. Moreover, a significant increase in the family Lactobacillaceae in IBS patients has been reported [73][74]. A review of the relationship between IBS and the gut microbiome revealed that the genera Faecalibacterium and Bifid
Recently, gut microbiome studies have shown greater abundances and higher gut richness of the Clostridia class, the families Ruminococcaceae and Prevotellaceae, and the Faecalibacterium and Roseburia genera in individual patients colonized with Blastocystis [46][75]. However, individuals not colonized with Blastocystis exhibited a higher abundance of Bacteroides [53].
3. Blastocystis and the Gut–Brain Axis
The communication between the brain and the gut microbiome is bidirectional and is termed the “gut microbiome–brain axis”. Communication along the gut–brain axis is mediated by various transmission systems, including the enteric nervous system, central nervous system, immune system, and endocrine system [76]. Maintaining a good balance between the gut microbiome and the brain is important for the host [77].
Although research has been conducted on the relationship between parasite manipulations and insect parasite interactions with the central nervous system (CNS), there have not been many studies on the interaction of the vertebrate host CNS and parasites [78][79]. The understanding of the interaction of the host CNS and parasites has increased recently with the development of the new and developing field of neuro-parasitology. Parasites can significantly affect the functioning of the host organism, including the immune response and the gut–brain axis, resulting in altered host behavior [80]. Echinococcus granulosus-derived ESPs (excretory–secretory products) affect cognitive function and the gut microbiome–brain axis as they have been demonstrated to alleviate dysbiosis and ameliorate cognitive decline in obese mice [81].
Despite the uncertainty surrounding the parasitic nature of Blastocystis, studies such as the above can shed light on the gut–brain axis relation to Blastocystis colonization/infection. There have been a limited number of studies showing the mechanisms through which the presence of Blastocystis in the intestine might influence the cognitive behavior of the host.
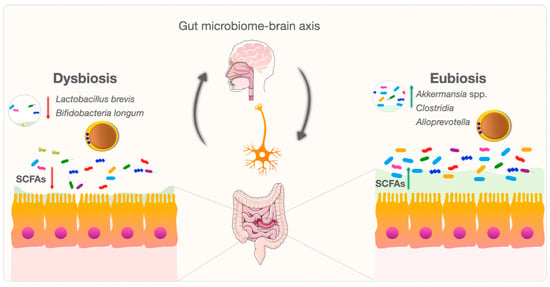
The gastrointestinal system is a complex and dynamic environment. Blastocystis exhibits broad genetic diversity, and the mechanisms and relationships between various subspecies and eubiosis/dysbiosis are being investigated [82][83][84][85]. Changes in gut microbiota species and critical metabolite levels in Blastocystis-colonized individuals may produce various potent signaling molecules in tryptophan metabolism [82][86]. These molecules may influence the gut microbiome–brain axis by altering tryptophan levels in gastrointestinal and neurological signaling pathways [82][87]. Blastocystis may also contribute to the balance of the bidirectional gut–brain axis (Figure 2). Blastocystis needs to be further considered as a new and mysterious actor in gut microbiome–brain axis research.

Figure 2. Bidirectional modulation and interaction of the gut microbiome–brain axis between Blastocystis and the gut microbiome.
4. Blastocystis and Probiotics
Probiotics are specific microorganisms that have beneficial effects on health. The most commonly used probiotics are specific strains from lactic acid bacterial species, especially Lactobacillus strains (Streptococcus thermophilus, Lactococcus lactis, Enterococcus faecium, and others) and Bifidobacterium strains and the yeast Saccharomyces boulardii (S. boulardii). Probiotics can modulate the microbiota and immune response of the host and inhibit the proliferation of parasites, leading to reduced parasitological loads and clinical improvement. Moreover, probiotics can increase the abundance of beneficial bacteria in the microbiota, change the environmental conditions to become less favorable for pathogens, compete with pathogens for nutrients and adhesion sites pathogens, negatively affect pathogens with their useful secretions (i.e., bacteriocins, lactic acid, hydrogen peroxide, etc.), inhibit bacterial toxins, increase mucus secretion, and induce mucosal immunity [88][89][90][91][92][93][94][95]. Although the relationship between probiotics and parasites has been investigated in various studies [88][92][96][97][98][99][100][101][102], there are very few reports related to Blastocystis and probiotics.
Blastocystis infections can occur in different forms ranging from asymptomatic to severe. Furthermore, the detection of Blastocystis in a stool sample does not necessarily mean that treatment is required. Its presence can be associated with infection or colonization whereby Blastocystis is a member of the healthy gut microbiome. This variability in outcomes could be due to different subtypes, the immunological response of the host, and gut microbial diversity [21][45][52][75][103][104][105][106]. If treatment is decided upon in the required symptomatic group (gastrointestinal symptoms; dermatological disorders involving acute/chronic urticaria and itching), the first choice is metronidazole.
Dinleyici et al. compared therapies with S. boulardii and metronidazole in symptomatic children with a Blastocystis infection. They assessed clinical and parasitological cures in both study groups. While both metronidazole and S. boulardii demonstrated potential beneficial effects in treating Blastocystis infection, no statistically significant difference was found between the two treatment groups [107].
The possible effects of probiotics on Blastocystis along with the type and dose of probiotic used for treatment remain unclear. In addition to the views that probiotics have treatment potential for Blastocystis, some studies argue that Blastocystis is a member of the healthy microbiota and that some Blastocystis subtypes may themselves be used as probiotics in the future [106]. This may also be an intriguing research subject. In the future, more successful results can be achieved with the use of probiotics designed by performing personalized microbiome analyses. Additional extensive studies are needed to achieve a comprehensive understanding.
This entry is adapted from the peer-reviewed paper 10.3390/microorganisms12030461
References
- Stensvold, C.R.; Tan, K.S.W.; Clark, C.G. Blastocystis. Trends Parasitol. 2020, 36, 315–316.
- Tan, K.S. New insights on classification, identification, and clinical relevance of Blastocystis spp. Clin. Microbiol. Rev. 2008, 21, 639–665.
- Popruk, S.; Pintong, A.-R.; Radomyos, P. Diversity of Blastocystis subtypes in humans. J. Trop. Med. Parasitol. 2013, 36, 88–97.
- El Safadi, D.; Cian, A.; Nourrisson, C.; Pereira, B.; Morelle, C.; Bastien, P.; Bellanger, A.-P.; Botterel, F.; Candolfi, E.; Desoubeaux, G. Prevalence, risk factors for infection and subtype distribution of the intestinal parasite Blastocystis sp. from a large-scale multi-center study in France. BMC Infect. Dis. 2016, 16, 451.
- Alfellani, M.A.; Jacob, A.S.; Perea, N.O.; Krecek, R.C.; Taner-Mulla, D.; Verweij, J.J.; Levecke, B.; Tannich, E.; Clark, C.G.; Stensvold, C.R. Diversity and distribution of Blastocystis sp. subtypes in non-human primates. Parasitology 2013, 140, 966–971.
- Santin, M.; Figueiredo, A.; Molokin, A.; George, N.S.; Köster, P.C.; Dashti, A.; González-Barrio, D.; Carmena, D.; Maloney, J.G. Division of Blastocystis ST10 into three new subtypes: ST42–ST44. J. Eukaryot. Microbiol. 2023, 71, e12998.
- Koehler, A.V.; Herath, H.D.; Hall, R.S.; Wilcox, S.; Gasser, R.B. Marked genetic diversity within Blastocystis in Australian wildlife revealed using a next generation sequencing–phylogenetic approach. Int. J. Parasitol. Parasites Wildl. 2024, 23, 100902.
- Stensvold, C.R.; Clark, C.G. Pre-empting Pandora’s Box: Blastocystis Subtypes Revisited. Trends Parasitol. 2020, 36, 229–232.
- Higuera, A.; Salas-Leiva, D.E.; Curtis, B.; Patiño, L.H.; Zhao, D.; Jerlström-Hultqvist, J.; Dlutek, M.; Muñoz, M.; Roger, A.J.; Ramírez, J.D. Draft genomes of Blastocystis subtypes from human samples of Colombia. Parasites Vectors 2023, 16, 52.
- Denoeud, F.; Roussel, M.; Noel, B.; Wawrzyniak, I.; Da Silva, C.; Diogon, M.; Viscogliosi, E.; Brochier-Armanet, C.; Couloux, A.; Poulain, J. Genome sequence of the stramenopile Blastocystis, a human anaerobic parasite. Genome Biol. 2011, 12, R29.
- Gentekaki, E.; Curtis, B.A.; Stairs, C.W.; Klimes, V.; Elias, M.; Salas-Leiva, D.E.; Herman, E.K.; Eme, L.; Arias, M.C.; Henrissat, B.; et al. Extreme genome diversity in the hyper-prevalent parasitic eukaryote Blastocystis. PLoS Biol. 2017, 15, e2003769.
- Robles-Cabrera, M.X.; Maguina, J.L.; Gonzales-Huerta, L.; Panduro-Correa, V.; Damaso-Mata, B.; Pecho-Silva, S.; Navarro-Solsol, A.C.; Rabaan, A.A.; Rodriguez-Morales, A.J.; Arteaga-Livias, K. Blastocystis species and Gastrointestinal Symptoms in Peruvian Adults Attended in a Public Hospital. Infect. Chemother. 2021, 53, 374–380.
- Cekin, A.H.; Cekin, Y.; Adakan, Y.; Tasdemir, E.; Koclar, F.G.; Yolcular, B.O. Blastocystosis in patients with gastrointestinal symptoms: A case–control study. BMC Gastroenterol. 2012, 12, 122.
- Coyle, C.M.; Varughese, J.; Weiss, L.M.; Tanowitz, H.B. Blastocystis: To treat or not to treat. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2012, 54, 105–110.
- Kurt, O.; Dogruman Al, F.; Tanyuksel, M. Eradication of Blastocystis in humans: Really necessary for all? Parasitol. Int. 2016, 65, 797–801.
- Bahrami, F.; Babaei, E.; Badirzadeh, A.; Riabi, T.R.; Abdoli, A. Blastocystis, urticaria, and skin disorders: Review of the current evidences. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 39, 1027–1042.
- Aykur, M.; Camyar, A.; Turk, B.G.; Sin, A.Z.; Dagci, H. Evaluation of association with subtypes and alleles of Blastocystis with chronic spontaneous urticaria. Acta Trop. 2022, 231, 106455.
- Nourrisson, C.; Wawrzyniak, I.; Cian, A.; Livrelli, V.; Viscogliosi, E.; Delbac, F.; Poirier, P. On Blastocystis secreted cysteine proteases: A legumain-activated cathepsin B increases paracellular permeability of intestinal Caco-2 cell monolayers. Parasitology 2016, 143, 1713–1722.
- Krogsgaard, L.R.; Engsbro, A.L.; Stensvold, C.R.; Nielsen, H.V.; Bytzer, P. The prevalence of intestinal parasites is not greater among individuals with irritable bowel syndrome: A population-based case-control study. Clin. Gastroenterol. Hepatol. 2015, 13, 507–513.e2.
- Petersen, A.M.; Stensvold, C.R.; Mirsepasi, H.; Engberg, J.; Friis-Moller, A.; Porsbo, L.J.; Hammerum, A.M.; Nordgaard-Lassen, I.; Nielsen, H.V.; Krogfelt, K.A. Active ulcerative colitis associated with low prevalence of Blastocystis and Dientamoeba fragilis infection. Scand. J. Gastroenterol. 2013, 48, 638–639.
- Scanlan, P.D.; Stensvold, C.R.; Rajilic-Stojanovic, M.; Heilig, H.G.; De Vos, W.M.; O’Toole, P.W.; Cotter, P.D. The microbial eukaryote Blastocystis is a prevalent and diverse member of the healthy human gut microbiota. FEMS Microbiol. Ecol. 2014, 90, 326–330.
- Parfrey, L.W.; Walters, W.A.; Lauber, C.L.; Clemente, J.C.; Berg-Lyons, D.; Teiling, C.; Kodira, C.; Mohiuddin, M.; Brunelle, J.; Driscoll, M.; et al. Communities of microbial eukaryotes in the mammalian gut within the context of environmental eukaryotic diversity. Front. Microbiol. 2014, 5, 298.
- Roberts, T.; Ellis, J.; Harkness, J.; Marriott, D.; Stark, D. Treatment failure in patients with chronic Blastocystis infection. J. Med. Microbiol. 2014, 63, 252–257.
- Cahana, I.; Iraqi, F.A. Impact of host genetics on gut microbiome: Take-home lessons from human and mouse studies. Anim. Model. Exp. Med. 2020, 3, 229–236.
- Gupta, V.K.; Paul, S.; Dutta, C. Geography, ethnicity or subsistence-specific variations in human microbiome composition and diversity. Front. Microbiol. 2017, 8, 1162.
- Kau, A.L.; Ahern, P.P.; Griffin, N.W.; Goodman, A.L.; Gordon, J.I. Human nutrition, the gut microbiome and the immune system. Nature 2011, 474, 327–336.
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227.
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836.
- Hall, A.B.; Tolonen, A.C.; Xavier, R.J. Human genetic variation and the gut microbiome in disease. Nat. Rev. Genet. 2017, 18, 690–699.
- Deidda, G.; Biazzo, M. Gut and Brain: Investigating Physiological and Pathological Interactions between Microbiota and Brain to Gain New Therapeutic Avenues for Brain Diseases. Front. Neurosci. 2021, 15, 753915.
- Vemuri, R.; Gundamaraju, R.; Shastri, M.D.; Shukla, S.D.; Kalpurath, K.; Ball, M.; Tristram, S.; Shankar, E.M.; Ahuja, K.; Eri, R. Gut Microbial Changes, Interactions, and Their Implications on Human Lifecycle: An Ageing Perspective. BioMed Res. Int. 2018, 2018, 4178607.
- Gevers, D.; Knight, R.; Petrosino, J.F.; Huang, K.; McGuire, A.L.; Birren, B.W.; Nelson, K.E.; White, O.; Methe, B.A.; Huttenhower, C. The Human Microbiome Project: A community resource for the healthy human microbiome. PLoS Biol. 2012, 10, e1001377.
- Ehrlich, S.D.; Consortium, M. MetaHIT: The European Union Project on Metagenomics of the Human Intestinal Tract. In Metagenomics of the Human Body; Springer: New York, NY, USA, 2011; pp. 307–316.
- Clarke, S.F.; Murphy, E.F.; O’Sullivan, O.; Lucey, A.J.; Humphreys, M.; Hogan, A.; Hayes, P.; O’Reilly, M.; Jeffery, I.B.; Wood-Martin, R. Exercise and associated dietary extremes impact on gut microbial diversity. Gut 2014, 63, 1913–1920.
- Jandhyala, S.M.; Talukdar, R.; Subramanyam, C.; Vuyyuru, H.; Sasikala, M.; Nageshwar Reddy, D. Role of the normal gut microbiota. World J. Gastroenterol. 2015, 21, 8787–8803.
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020, 30, 492–506.
- Hills, R.D., Jr.; Pontefract, B.A.; Mishcon, H.R.; Black, C.A.; Sutton, S.C.; Theberge, C.R. Gut Microbiome: Profound Implications for Diet and Disease. Nutrients 2019, 11, 1613.
- Zubeldia-Varela, E.; Barker-Tejeda, T.C.; Obeso, D.; Villasenor, A.; Barber, D.; Perez-Gordo, M. Microbiome and Allergy: New Insights and Perspectives. J. Investig. Allergol. Clin. Immunol. 2022, 32, 327–344.
- Gülden, E.; Wong, F.S.; Wen, L. The gut microbiota and type 1 diabetes. Clin. Immunol. 2015, 159, 143–153.
- Jalanka-Tuovinen, J.; Salojärvi, J.; Salonen, A.; Immonen, O.; Garsed, K.; Kelly, F.M.; Zaitoun, A.; Palva, A.; Spiller, R.C.; De Vos, W.M. Faecal microbiota composition and host–microbe cross-talk following gastroenteritis and in postinfectious irritable bowel syndrome. Gut 2014, 63, 1737–1745.
- Perry, R.J.; Peng, L.; Barry, N.A.; Cline, G.W.; Zhang, D.; Cardone, R.L.; Petersen, K.F.; Kibbey, R.G.; Goodman, A.L.; Shulman, G.I. Acetate mediates a microbiome–brain–β-cell axis to promote metabolic syndrome. Nature 2016, 534, 213–217.
- Vijay, A.; Valdes, A.M. Role of the gut microbiome in chronic diseases: A narrative review. Eur. J. Clin. Nutr. 2022, 76, 489–501.
- Thompson, R.; Ash, A. Molecular epidemiology of Giardia and Cryptosporidium infections. Infect. Genet. Evol. 2016, 40, 315–323.
- Dubik, M.; Pilecki, B.; Moeller, J.B. Commensal Intestinal Protozoa—Underestimated Members of the Gut Microbial Community. Biology 2022, 11, 1742.
- Billy, V.; Lhotska, Z.; Jirku, M.; Kadlecova, O.; Frgelecova, L.; Parfrey, L.W.; Pomajbikova, K.J. Blastocystis Colonization Alters the Gut Microbiome and, in Some Cases, Promotes Faster Recovery from Induced Colitis. Front. Microbiol. 2021, 12, 641483.
- Deng, L.; Wojciech, L.; Gascoigne, N.R.J.; Peng, G.; Tan, K.S.W. New insights into the interactions between Blastocystis, the gut microbiota, and host immunity. PLoS Pathog. 2021, 17, e1009253.
- Behboud, S.; Solhjoo, K.; Erfanian, S.; Pirestani, M.; Abdoli, A. Alteration of gut bacteria composition among individuals with asymptomatic Blastocystis infection: A case-control study. Microb. Pathog. 2022, 169, 105639.
- Nieves-Ramirez, M.E.; Partida-Rodriguez, O.; Laforest-Lapointe, I.; Reynolds, L.A.; Brown, E.M.; Valdez-Salazar, A.; Moran-Silva, P.; Rojas-Velazquez, L.; Morien, E.; Parfrey, L.W.; et al. Asymptomatic Intestinal Colonization with Protist Blastocystis Is Strongly Associated with Distinct Microbiome Ecological Patterns. mSystems 2018, 3.
- Asghari, A.; Hassanipour, S.; Hatam, G. Comparative molecular prevalence and subtypes distribution of Blastocystis sp. a potentially zoonotic infection isolated from symptomatic and asymptomatic patients in Iran: A systematic review and meta-analysis. Acta Parasitol. 2021, 66, 745–759.
- Ramirez, J.D.; Sanchez, A.; Hernandez, C.; Florez, C.; Bernal, M.C.; Giraldo, J.C.; Reyes, P.; Lopez, M.C.; Garcia, L.; Cooper, P.J.; et al. Geographic distribution of human Blastocystis subtypes in South America. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2016, 41, 32–35.
- Yowang, A.; Tsaousis, A.D.; Chumphonsuk, T.; Thongsin, N.; Kullawong, N.; Popluechai, S.; Gentekaki, E. High diversity of Blastocystis subtypes isolated from asymptomatic adults living in Chiang Rai, Thailand. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2018, 65, 270–275.
- Deng, L.; Wojciech, L.; Png, C.W.; Kioh, Y.Q.D.; Ng, G.C.; Chan, E.C.Y.; Zhang, Y.; Gascoigne, N.R.J.; Tan, K.S.W. Colonization with ubiquitous protist Blastocystis ST1 ameliorates DSS-induced colitis and promotes beneficial microbiota and immune outcomes. NPJ Biofilms Microbiomes 2023, 9, 22.
- Andersen, L.O.; Bonde, I.; Nielsen, H.B.; Stensvold, C.R. A retrospective metagenomics approach to studying Blastocystis. FEMS Microbiol. Ecol. 2015, 91, fiv072.
- Deng, L.; Wojciech, L.; Png, C.W.; Kioh, D.Y.Q.; Gu, Y.; Aung, T.T.; Malleret, B.; Chan, E.C.Y.; Peng, G.; Zhang, Y. Colonization with two different Blastocystis subtypes in DSS-induced colitis mice is associated with strikingly different microbiome and pathological features. Theranostics 2023, 13, 1165.
- Gallo, A.; Passaro, G.; Gasbarrini, A.; Landolfi, R.; Montalto, M. Modulation of microbiota as treatment for intestinal inflammatory disorders: An uptodate. World J. Gastroenterol. 2016, 22, 7186–7202.
- Deng, L.; Wojciech, L.; Png, C.W.; Koh, E.Y.; Aung, T.T.; Kioh, D.Y.Q.; Chan, E.C.Y.; Malleret, B.; Zhang, Y.; Peng, G.; et al. Experimental colonization with Blastocystis ST4 is associated with protective immune responses and modulation of gut microbiome in a DSS-induced colitis mouse model. Cell. Mol. Life Sci. 2022, 79, 245.
- Rojas-Velázquez, L.; Morán, P.; Serrano-Vázquez, A.; Portillo-Bobadilla, T.; González, E.; Pérez-Juárez, H.; Hernández, E.; Partida-Rodríguez, O.; Nieves-Ramírez, M.; Padilla, A.; et al. The regulatory function of Blastocystis spp. on the immune inflammatory response in the gut microbiome. Front. Cell. Infect. Microbiol. 2022, 12, 967724.
- Betts, E.L.; Newton, J.M.; Thompson, G.S.; Sarzhanov, F.; Jinatham, V.; Kim, M.-J.; Popluechai, S.; Dogruman-Al, F.; Won, E.-J.; Gentekaki, E. Metabolic fluctuations in the human stool obtained from Blastocystis carriers and non-carriers. Metabolites 2021, 11, 883.
- Tan, K.S.W.; Mirza, H. Blastocystis–Host Interactions: Insights from In Vitro Model Systems. In Blastocystis: Pathogen or Passenger? An Evaluation of 101 Years of Research; Mehlhorn, H., Tan, K.S.W., Yoshikawa, H., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 51–63.
- Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, A.T.; Creasy, H.H.; Earl, A.M.; FitzGerald, M.G.; Fulton, R.S.; et al. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214.
- Ahrodia, T.; Das, S.; Bakshi, S.; Das, B. Structure, functions, and diversity of the healthy human microbiome. Prog. Mol. Biol. Transl. 2022, 191, 53–82.
- Belzer, C.; Chia, L.W.; Aalvink, S.; Chamlagain, B.; Piironen, V.; Knol, J.; de Vos, W.M. Microbial metabolic networks at the mucus layer lead to diet-independent butyrate and vitamin B12 production by intestinal symbionts. Mbio 2017, 8, e00770-17.
- Llinás-Caballero, K.; Caraballo, L. Helminths and bacterial microbiota: The interactions of two of humans’ “old friends”. Int. J. Mol. Sci. 2022, 23, 13358.
- Woof, J.M.; Kerr, M.A. The function of immunoglobulin A in immunity. J. Pathol. J. Pathol. Soc. Great Br. Irel. 2006, 208, 270–282.
- Puthia, M.K.; Vaithilingam, A.; Lu, J.; Tan, K.S. Degradation of human secretory immunoglobulin A by Blastocystis. Parasitol. Res. 2005, 97, 386–389.
- Puthia, M.K.; Lu, J.; Tan, K.S. Blastocystis ratti contains cysteine proteases that mediate interleukin-8 response from human intestinal epithelial cells in an NF-κB-dependent manner. Eukaryot. Cell 2008, 7, 435–443.
- Shaheen, W.A.; Quraishi, M.N.; Iqbal, T.H. Gut microbiome and autoimmune disorders. Clin. Exp. Immunol. 2022, 209, 161–174.
- De Luca, F.; Shoenfeld, Y. The microbiome in autoimmune diseases. Clin. Exp. Immunol. 2019, 195, 74–85.
- Bakland, G.; Nossent, H.C. Epidemiology of spondyloarthritis: A review. Curr. Rheumatol. Rep. 2013, 15, 351.
- Stoll, M.L.; Weiss, P.F.; Weiss, J.E.; Nigrovic, P.A.; Edelheit, B.S.; Bridges, S.L., Jr.; Danila, M.I.; Spencer, C.H.; Punaro, M.G.; Schikler, K.; et al. Age and fecal microbial strain-specific differences in patients with spondyloarthritis. Arthritis Res. Ther. 2018, 20, 14.
- Wang, L.; Wang, Y.; Zhang, P.; Song, C.; Pan, F.; Li, G.; Peng, L.; Yang, Y.; Wei, Z.; Huang, F. Gut microbiota changes in patients with spondyloarthritis: A systematic review. Semin. Arthritis Rheum. 2022, 52, 151925.
- Shaikh, S.D.; Sun, N.; Canakis, A.; Park, W.Y.; Weber, H.C. Irritable Bowel Syndrome and the Gut Microbiome: A Comprehensive Review. J. Clin. Med. 2023, 12, 2558.
- Chung, C.S.; Chang, P.F.; Liao, C.H.; Lee, T.H.; Chen, Y.; Lee, Y.C.; Wu, M.S.; Wang, H.P.; Ni, Y.H. Differences of microbiota in small bowel and faeces between irritable bowel syndrome patients and healthy subjects. Scand. J. Gastroenterol. 2016, 51, 410–419.
- Carroll, I.M.; Ringel-Kulka, T.; Siddle, J.P.; Ringel, Y. Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol. Motil. 2012, 24, 521-e248.
- Audebert, C.; Even, G.; Cian, A.; The Blastocystis Investigation Group; Loywick, A.; Merlin, S.; Viscogliosi, E.; Chabe, M. Colonization with the enteric protozoa Blastocystis is associated with increased diversity of human gut bacterial microbiota. Sci. Rep. 2016, 6, 25255.
- Liu, L.; Huh, J.R.; Shah, K. Microbiota and the gut-brain-axis: Implications for new therapeutic design in the CNS. EBioMedicine 2022, 77, 103908.
- Hillestad, E.M.R.; van der Meeren, A.; Nagaraja, B.H.; Bjorsvik, B.R.; Haleem, N.; Benitez-Paez, A.; Sanz, Y.; Hausken, T.; Lied, G.A.; Lundervold, A.; et al. Gut bless you: The microbiota-gut-brain axis in irritable bowel syndrome. World J. Gastroenterol. 2022, 28, 412–431.
- Blecharz-Klin, K.; Swierczynska, M.; Piechal, A.; Wawer, A.; Joniec-Maciejak, I.; Pyrzanowska, J.; Wojnar, E.; Zawistowska-Deniziak, A.; Sulima-Celinska, A.; Mlocicki, D.; et al. Infection with intestinal helminth (Hymenolepis diminuta) impacts exploratory behavior and cognitive processes in rats by changing the central level of neurotransmitters. PLoS Pathog. 2022, 18, e1010330.
- Carabotti, M.; Scirocco, A.; Maselli, M.A.; Severi, C. The gut-brain axis: Interactions between enteric microbiota, central and enteric nervous systems. Ann. Gastroenterol. 2015, 28, 203–209.
- McKenney, E.A.; Williamson, L.; Yoder, A.D.; Rawls, J.F.; Bilbo, S.D.; Parker, W. Alteration of the rat cecal microbiome during colonization with the helminth Hymenolepis diminuta. Gut Microbes 2015, 6, 182–193.
- Wu, J.; Zhu, Y.; Zhou, L.; Lu, Y.; Feng, T.; Dai, M.; Liu, J.; Xu, W.; Cheng, W.; Sun, F.; et al. Parasite-Derived Excretory-Secretory Products Alleviate Gut Microbiota Dysbiosis and Improve Cognitive Impairment Induced by a High-Fat Diet. Front. Immunol. 2021, 12, 710513.
- Leonardi, S.S.; Tan, K.S.-W. Blastocystis: View from atop the gut–brain iceberg. Trends Parasitol. 2023, 40, 1–4.
- Rieder, R.; Wisniewski, P.J.; Alderman, B.L.; Campbell, S.C. Microbes and mental health: A review. Brain Behav. Immun. 2017, 66, 9–17.
- Khine, W.W.T.; Voong, M.L.; Ng, T.K.S.; Feng, L.; Rane, G.A.; Kumar, A.P.; Kua, E.H.; Mahendran, R.; Mahendran, R.; Lee, Y.K. Mental awareness improved mild cognitive impairment and modulated gut microbiome. Aging 2020, 12, 24371–24393.
- Stensvold, C.R.; van der Giezen, M. Associations between Gut Microbiota and Common Luminal Intestinal Parasites. Trends Parasitol. 2018, 34, 369–377.
- Correia, A.S.; Vale, N. Tryptophan metabolism in depression: A narrative review with a focus on serotonin and kynurenine pathways. Int. J. Mol. Sci. 2022, 23, 8493.
- Nozawa, K.; Kawabata-Shoda, E.; Doihara, H.; Kojima, R.; Okada, H.; Mochizuki, S.; Sano, Y.; Inamura, K.; Matsushime, H.; Koizumi, T. TRPA1 regulates gastrointestinal motility through serotonin release from enterochromaffin cells. Proc. Natl. Acad. Sci. USA 2009, 106, 3408–3413.
- Travers, M.-A.; Florent, I.; Kohl, L.; Grellier, P. Probiotics for the control of parasites: An overview. J. Parasitol. Res. 2011, 2011, 610769.
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514.
- Vitetta, L.; Saltzman, E.T.; Nikov, T.; Ibrahim, I.; Hall, S. Modulating the gut micro-environment in the treatment of intestinal parasites. J. Clin. Med. 2016, 5, 102.
- Vallianou, N.; Stratigou, T.; Christodoulatos, G.S.; Tsigalou, C.; Dalamaga, M. Probiotics, Prebiotics, Synbiotics, Postbiotics, and Obesity: Current Evidence, Controversies, and Perspectives. Curr. Obes. Rep. 2020, 9, 179–192.
- Sarid, L.; Zanditenas, E.; Ye, J.; Trebicz-Geffen, M.; Ankri, S. Insights into the mechanisms of Lactobacillus acidophilus activity against Entamoeba histolytica by using thiol redox proteomics. Antioxidants 2022, 11, 814.
- Hardy, H.; Harris, J.; Lyon, E.; Beal, J.; Foey, A.D. Probiotics, prebiotics and immunomodulation of gut mucosal defences: Homeostasis and immunopathology. Nutrients 2013, 5, 1869–1912.
- Markowiak, P.; Śliżewska, K. Effects of probiotics, prebiotics, and synbiotics on human health. Nutrients 2017, 9, 1021.
- Silva, D.R.; Sardi, J.d.C.O.; de Souza Pitangui, N.; Roque, S.M.; da Silva, A.C.B.; Rosalen, P.L. Probiotics as an alternative antimicrobial therapy: Current reality and future directions. J. Funct. Foods 2020, 73, 104080.
- Berrilli, F.; Di Cave, D.; Cavallero, S.; D’Amelio, S. Interactions between parasites and microbial communities in the human gut. Front. Cell. Infect. Microbiol. 2012, 2, 141.
- Pryshliak, O.Y.; Protsyk, A.L.; Semaniv, M.V.; Boichuk, O.P.; Gerych, P.R. Effect of probiotics on the intestinal microbiota of patients with giardiasis and ascariasis. J. Med. Life 2022, 15, 1278.
- Al-Megrin, W.A.; Mohamed, S.H.; Saleh, M.M.; Yehia, H.M. Preventive role of probiotic bacteria against gastrointestinal diseases in mice caused by Giardia lamblia. Biosci. Rep. 2021, 41, BSR20204114.
- Del Coco, V.F.; Sparo, M.D.; Sidoti, A.; Santín, M.; Basualdo, J.A.; Córdoba, M.A. Effects of Enterococcus faecalis CECT 7121 on Cryptosporidium parvum infection in mice. Parasitol. Res. 2016, 115, 3239–3244.
- Shukla, G.; Sharma, A.; Bhatia, R.; Sharma, M. Prophylactic potential of synbiotic (Lactobacillus casei and Inulin) in malnourished murine giardiasis: An immunological and ultrastructural study. Probiotics Antimicrob. Proteins 2019, 11, 165–174.
- Saracino, M.P.; Vila, C.C.; Baldi, P.C.; Gonzalez Maglio, D.H. Searching for the one (s): Using probiotics as anthelmintic treatments. Front. Pharmacol. 2021, 12, 714198.
- Rooney, J.; Cantacessi, C.; Sotillo, J.; Cortés, A. Gastrointestinal worms and bacteria: From association to intervention. Parasite Immunol. 2023, 45, e12955.
- Beghini, F.; Pasolli, E.; Truong, T.D.; Putignani, L.; Caccio, S.M.; Segata, N. Large-scale comparative metagenomics of Blastocystis, a common member of the human gut microbiome. ISME J. 2017, 11, 2848–2863.
- Yason, J.A.; Liang, Y.R.; Png, C.W.; Zhang, Y.; Tan, K.S.W. Interactions between a pathogenic Blastocystis subtype and gut microbiota: In vitro and in vivo studies. Microbiome 2019, 7, 30.
- Deng, L.; Tan, K.S.W. Interactions between Blastocystis subtype ST4 and gut microbiota in vitro. Parasites Vectors 2022, 15, 80.
- Andersen, L.O.; Stensvold, C.R. Blastocystis in Health and Disease: Are We Moving from a Clinical to a Public Health Perspective? J. Clin. Microbiol. 2016, 54, 524–528.
- Dinleyici, E.C.; Eren, M.; Dogan, N.; Reyhanioglu, S.; Yargic, Z.A.; Vandenplas, Y. Clinical efficacy of Saccharomyces boulardii or metronidazole in symptomatic children with Blastocystis hominis infection. Parasitol. Res. 2011, 108, 541–545.
This entry is offline, you can click here to edit this entry!
