Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Heart failure (HF) is a widespread cardiovascular condition that poses significant risks to a wide spectrum of age groups and leads to terminal illness. Although our understanding of the underlying mechanisms of HF has improved, the available treatments still remain inadequate. Recently, long non-coding RNAs (lncRNAs) have emerged as crucial players in cardiac function, showing possibilities as potential targets for HF therapy. These versatile molecules interact with chromatin, proteins, RNA, and DNA, influencing gene regulation.
- long non-coding RNAs (lncRNAs)
- heart failure
- diagnostic markers
- therapeutic targets
- gene regulation
- cardiovascular biomarkers
1. Introduction
Heart failure (HF) is a complex and impairing cardiovascular disease that affects millions of individuals worldwide, causing increasing rates of morbidity and mortality [1]. The molecular mechanisms underlying HF pathogenesis are multifaceted and involve the dysregulation of various genes and signaling pathways. Recent research has shed light on the crucial role of lncRNAs in modulating gene expression and cellular processes, making them prominent players in the context of HF [2][3].
Long non-coding RNAs (lncRNAs) constitute a diverse group of RNA molecules with lengths surpassing 200 nucleotides and lacking protein-coding abilities [4]. Initially disregarded as mere transcriptional background noise, lncRNAs have now emerged as pivotal regulators of gene expression and cellular processes. They exercise their impact via various mechanisms, including chromatin remodeling, transcriptional interference, and post-transcriptional changes [5][6][7]. Their involvement in regulating essential pathways within cardiovascular biology, such as apoptosis, inflammation, and the remodeling of cardiac tissues, are also highly noteworthy [8][9].
2. Understanding Heart Failure: A Brief Overview
HF is a complex cardiovascular disorder where the heart struggles to pump blood efficiently, leading to inadequate supply to vital organs [10][11]. It is essential to grasp the complexities of HF for effectively managing this condition, given its significant impact on clinical outcomes (Figure S1, can be downloaded at: https://www.mdpi.com/article/10.3390/ncrna10010003/s1). To truly understand how HF develops, we need to consider its multifaceted nature. This involves changes in both the structure and function of the heart, as well as the activation of certain hormonal pathways and inflammatory response.
As HF progresses, we typically see enlargement of heart cells, the development of fibrous tissue, compromised heart muscle contractility, and the heart undergoing maladaptive changes. Additionally, hormonal responses like the renin–angiotensin–aldosterone system (RAAS) and the sympathetic nervous system (SNS) kick in due to reduced heart output, leading to blood vessel constriction and the retention of sodium and water. These factors further worsen HF. Therefore, gaining a deep understanding of how these various processes interact is essential for creating effective approaches for preventing HF, diagnosing it early, and providing proper management [12][13].
Figure 1 and Figure S1 show a schematic illustration of some studies in vitro and in vivo on the study of regulatory activity of lncRNAs in HF. The activity of lncRNAs in HF is based on a fine line of regulation of complex molecular networks at the level of epigenetic changes, transcription, and post-transcriptional transformations, such as lncRNA/microRNA (miRNA)/mRNA connections, which are necessary for the control of proliferation and migration of cardiomyocytes, collagen synthesis, inflammation, atrial electrical remodeling, cardiac autonomic nervous function, and oxidative stress.
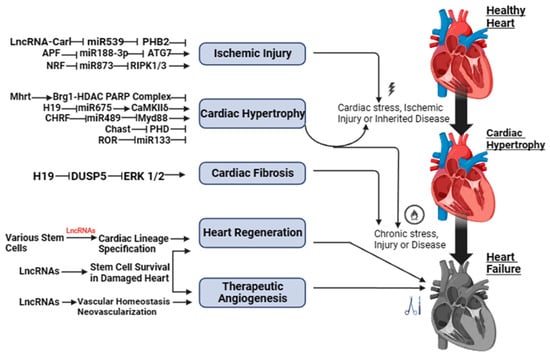
Figure 1. LncRNAs as possible therapeutic targets for heart failure.
3. Exploring the Connection between lncRNA and Heart Failure
Lately, the complex connection between lncRNAs and HF has become a focal point in cardiovascular research. This relationship holds the promise of unveiling novel insights into the molecular underpinnings of HF and, consequently, transforming our approach to diagnosis and treatment of this prevalent and debilitating condition [4][14][15].
Transpiring studies have brought to light the pivotal roles played by lncRNAs in the pathogenesis of HF [16]. Greco et al. [17] demonstrated that lncRNAs are intimately involved in orchestrating processes such as cardiomyocyte hypertrophy, fibrosis, apoptosis, and contractility, collectively contributing to the development and progression of HF [18]. These regulatory molecules, once dismissed as transcriptional noise, are now recognized as central players in the complex web of molecular interactions driving HF pathophysiology. The ever-evolving high-throughput RNA sequencing technologies have allowed researchers to identify dysregulated lncRNAs in HF, both in animal models and in humans [19][20]. All of these findings propose that lncRNAs play an explicative role in the pathogenesis of, and may pose as a potential therapeutic target, for this life-threatening condition [21][22]. One of the remarkable aspects of the lncRNA–HF connection is the diversity of regulatory mechanisms involved [23][24]; for example, the lncRNA H19 has been shown to promote myocardial fibrosis by influencing the deposition of extracellular matrix components in the heart. H19 is a long non-coding RNA that has been associated with various biological processes, including cellular growth, differentiation, and apoptosis. Several articles suggest that lncRNA-H19 is a potent regulator of the progression of cardiac hypertrophy. They indicate that lncRNA-H19 is involved in the pathophysiological processes of cardiac hypertrophy, including calcium regulation, fibrosis, apoptosis, angiogenesis, inflammation, and methylation [25][26]. In contrast, the lncRNA Mhrt (Myosin Heavy Chain Associated RNA Transcripts) has been found to protect against HF by inhibiting cardiac hypertrophy [27]. These findings underscore the complex and context-dependent roles of lncRNAs in HF.
Clinical Evidence
Recent clinical evidence shows that particular lncRNAs in HF can diagnose and give prognosis, possibly changing risk assessment and patient care. Li et al. and Kumarswamy et al. [28][29] identified lncRNAs, including ANRIL (Antisense Non-coding RNA in the INK4 Locus) and LIPCAR (mitochondrial long non-coding RNA uc022bqs.1), as prospective biomarkers for HF [30] due to their altered expression patterns in HF patients and their correlation with disease severity and prognosis. Their diagnostic and prognostic capabilities enable more accurate risk stratification and early HF identification, improving patient care [31].
The quantity and cellular origins of circulating lncRNAs in HF and their association with cardiac regeneration need more study [21][24]. The preliminary findings herein shed light on lncRNAs and HF. The preliminary data imply that lncRNAs are increasingly relevant in coronary heart disease, heart failure, and hypertension [32]. Due to their modest number and unknown cell source, circulating lncRNAs are difficult to detect. The link between circulating lncRNAs and cardiac regeneration is unclear, reducing the sensitivity and specificity of employing them as heart failure diagnostic markers or therapeutic targets [21][33].
Therefore, we must acknowledge the uncertainties and challenges in lncRNA research, which is crucial for a comprehensive understanding. It is evident that while lncRNAs hold immense potential in deciphering heart-related conditions, navigating through these complexities demands rigorous evaluation and standardized methodologies. Emphasizing the need for stringent evaluation and standardized methodologies is pivotal. Because of their complexity, lncRNAs must be thoroughly investigated and their roles and interactions must be validated using strong methodologies [24]. Standardization across research methodologies and data analysis frameworks would enhance comparability and reliability among studies, fostering a more cohesive understanding of lncRNA functions in heart failure and related conditions [32]. This rigorous approach is essential for establishing concrete correlations between specific lncRNAs and their functional implications in heart failure. It will also ensure that findings are reproducible and applicable in clinical settings [21]. Moreover, lncRNA-targeted therapeutics must be systematically examined in preclinical and clinical studies, and the possible off-target effects and long-term impacts of modifying lncRNAs should be carefully considered at every stage [34][35].
4. Unraveling the Role of lncRNAs in Heart Disease Pathogenesis
According to present-day knowledge, lncRNAs have emerged as crucial players in various biological processes, including the development and maintenance of heart function [36][37]. Through extensive research and advances in RNA sequencing technology, it has been discovered that LncRNAs exhibit distinct expression patterns in human heart failure compared with normal donor hearts [19][38]. Hence, it could be said that LncRNAs may play a regulatory role in the pathogenesis of heart failure [14][39]. Several studies have also reported the detection of lncRNAs in plasma or urine samples, which indicates their potential as diagnostic markers for HF [40].
In the context of HF, lncRNAs have been linked to critical cellular processes such as cardiomyocyte hypertrophy, fibrosis, apoptosis, and contractility [17][35]. These regulatory roles of lncRNAs have important implications for the development and progression of HF [41]. For example, antisense lncRNA Kcna2 (Long non-coding RNA Potassium Voltage-Gated Channel Subfamily A Member 2) is linked to a higher prevalence of ventricular arrhythmias, which are irregular heartbeats that only impact the heart’s lower chambers in heart failure patients [42]. The study found that Kcna2 knockdown in the heart decreased the slow component of the rectifier potassium current (IKs) and prolonged action potentials in cardiomyocytes, consistent with the changes observed in heart failure. Conversely, Kcna2 overexpression in the heart significantly attenuated the CHF-induced decreases in the IKs, AP prolongation, and ventricular arrhythmias. This discovery highlights the role of lncRNAs in cardiac electrical remodeling and the development of arrhythmias in heart failure [42]. Additionally, lncRNA UCA1 (Long non-coding RNA urothelial carcinoma-associated 1) has been shown to promote the progression of cardiac hypertrophy, a condition commonly associated with various cardiovascular diseases such as heart failure. Cardiac hypertrophy is closely associated with a series of cardiovascular diseases, including heart failure and sudden death in particular. Therefore, understanding the pathogenesis of cardiac hypertrophy is crucial for improving its diagnosis and therapy. According to a study, lncRNA UCA1 is a novel regulator in cardiomyocyte hypertrophy through targeting the miR-184/HOXA9 axis [43]. The study found that lncRNA UCA1 was highly expressed in mice heart treated with transverse aortic constriction (TAC) and the cardiomyocytes treated with phenylephrine (PE). On the contrary, miR-184 was downregulated under the same conditions. The knockdown of UCA1 or the overexpression of miR-184 lessened the enlarged surface area of cardiomyocytes and the elevated expressions of fetal genes (ANP and BNP) induced by PE. Later, it was determined that miR-184 was a direct target of UCA1, whereas the mRNA HOXA9 was a target of miR-184. Rescue assays indicated that UCA1 promoted the progression of cardiac hypertrophy through competitively binding with miR-184 to enhance the expression of HOXA9 [43].
Moreover, lncRNA GAS5 (Long non-coding RNA Growth Arrest-Specific 5), which is known as a tumor suppressor gene, has been implicated in heart failure caused by myocardial infarction. GAS5 is a long non-coding RNA that is a member of the 5′ terminal oligo-pyrimidine class of genes. It is a small nucleolar RNA host gene, containing multiple C/D box snoRNA genes in its introns. The secondary RNA structure of the encoded transcript mimics glucocorticoid response element (GRE), which means it can bind to the DNA binding domain of the glucocorticoid receptor (GR) and block its activation, thereby stopping it from regulating the transcription of its target genes [42]. GAS5 has been linked to apoptosis and cellular growth arrest. It may also function as a tumor suppressor; in several tissues, its downregulation has been linked to cancer. Several lncRNAs, including GAS5, are crucial regulators of cell differentiation, development, and disease, and their expression is often associated with stress conditions in the heart. The dysregulation of lncRNAs is associated with a variety of cardiac diseases, including heart failure [44]. However, the specific role of GAS5 in heart failure is yet to be determined. To study the same, a study was conducted by Wang and Xie [42] that aimed to evaluate the importance and its role in heart failure development post-myocardial infarction of lncRNA GAS5 in hypoxia-injured H9c2 cells, which is a type of cardiac muscle cell line that was derived from embryonic rat hearts. The results of this study demonstrated that lncRNA GAS5 was significantly upregulated in hypoxia-injured H9c2 cells and its expression inhibited the progression of heart failure caused by myocardial infarction [42]. It is also known that LncRNAs take part in interactions with several major regulators that are involved in the modulation of calcium ions, such as zinc finger antisense 1 (ZFAS1), myocardial infarction-associated transcript (Miat), and zinc finger protein antisense RNA (ZNF593-AS) [45][46]. Theoretically, the interaction between ZFAS1 and SERCA2a (Sarco/Endoplasmic Reticulum Calcium ATPase 2a, a crucial protein found in the sarcoplasmic reticulum membranes of cardiac muscle cells that controls calcium ion transport and heart muscle contraction and relaxation) leads to an excessive buildup of intracellular calcium ions, which causes abnormal calcium ion fluctuations in cardiomyocytes and impairs their ability to contract effectively. Miat has been observed to interfere with the pan-RNA splicing process, resulting in a reduction in the expression levels of SERCA2a and RyR2 (Ryanodine Receptor 2, a calcium channel mostly present in the cardiac muscle cells’ sarcoplasmic reticulum that is crucial for regulating the release of calcium during muscle contraction, particularly in the heart) [45][46]. Consequently, this disruption contributes to compromised contractility.
In addition to that, in a study conducted by Jiao et al., it was observed that lncRNA-ZFAS1 triggers mitochondria-mediated apoptosis in mice with myocardial infarction by inducing an excessive accumulation of cytosolic Ca2+ [47]. The reversible nature of the effect of ZFAS1 was observed following the knockdown, suggesting that targeting ZFAS1 with anti-ZFAS1 agents could serve as a novel therapeutic approach to safeguard cardiomyocytes from apoptosis generated by myocardial infarction [48][49]. Additionally, the LncRNA Caren (cardiomyocyte-enriched non-coding transcript), which is highly expressed in cardiomyocytes, plays a vital role in preserving cardiac function when pressure is high. It achieves this by suppressing the ataxia telangiectasia mutated (ATM)/DNA damage response (DDR) pathway and modulating the deacetylase activity of sirtuin 2 [50]. Another lncRNA, known as H19, shows a high degree of conservation and serves as a critical factor in the early stages of postnatal development as well as in the pathogenesis of several disorders [9][51][52]. Similarly, the expression of LncKCND1 (potassium voltage-gated channel subfamily D member 1) is increased in models of heart failure, and it functions to suppress the enlargement of cardiomyocytes by the production of miR-675 [53]. This microRNA specifically targets CaMKIIδ (Calcium/Calmodulin-Dependent Protein Kinase II delta, an enzyme that plays an important part in a variety of physiological activities, including calcium control and the formation of memory in neurons, as well as heart muscle contraction). Other lncRNAs involved in epigenetic regulation, such as the cardiac-hypertrophy-associated epigenetic regulator (CHAER), have direct interaction with PRC2 (Polycomb Repressive Complex 2, a protein complex involved in gene regulation control) [54]. This interaction does not allow PRC2 to target certain genomic areas, leading to a decrease in H3K27me3 (Histone 3 Lysine 27 trimethylation, a specific epigenetic modification that is associated with gene repression and in the regulation of chromatin structure and gene expression) levels at the promoter regions of genes implicated in cardiac hypertrophy [55].
LncRNAs have a substantial influence on the progression of cardiac fibrosis and heart failure due to myocardial infarction [56]. These molecules possess the ability to regulate the expression of neighboring genes, promote the growth and viability of fibroblasts, and impede the interaction between COTL1 (Coactosin-like protein 1, a protein involved in the regulation of actin filament dynamics) and TRAP1 (TNF receptor-associated protein 1, a member of the HSP 90- Heat Shock Protein 90 family and is found majorly in the mitochondria), allowing the transdifferentiation of myofibroblasts into cardiac fibroblasts [57].
Some exceptional work conducted by Han et al. investigated the expression patterns of lncRNA in human cardiac fibroblasts (HCFs) affected by cardiac fibrosis. The results of the functional analysis demonstrated that a total of 176 lncRNAs were elevated, while 526 lncRNAs were downregulated in human dermal fibroblasts (HCFs) that were stimulated with transforming growth factor-beta (TGF-β). The identified target genes were shown to be associated with several biological processes, including focal adhesion, metabolic pathways, the Hippo signaling system, the PI3K-Akt (Phosphoinositide 3-kinase-Protein Kinase B) signaling pathway, control of actin cytoskeleton, and hypertrophic cardiomyopathy [18][58][59]. The novel lncRNAs identified as NONHSAG005537 and NONHSAG017620 were also found to exert inhibitory effects on the proliferation, migration, invasion, and transformation of human cardiac fibroblasts (HCFs).
The CANTOS (Canakinumab Anti-inflammatory Thrombosis Outcome Study) trial also provides evidence supporting the potential efficacy of anticytokine therapy as a strategy for enhancing heart function in patients with myocardial infarction. Nevertheless, there is a significant lack of understanding regarding the specific mechanisms by which lncRNAs contribute to the development of cardiac dysfunction. However, according to a recent study, it has been discovered that the initiation of heart failure is facilitated by the action of SOX2-OT (SOX2 Overlapping Transcript, an lncRNA encoding factors that are essential to embryonic development and also involved in cell fate determination) through its interaction with miR-455-3p, increasing the expression of TNF receptor-associated factor 6 (TRAF6) and the subsequent activation of the NF-κb (Nuclear Factor kappa-light-chain-enhancer of activated B cells, a protein complex that regulates the expression of immune response genes) signaling pathway [60][61]. A translational study was conducted to investigate the role of lncRNAs in the antiviral response of individuals with coxsackievirus-B3 cardiomyopathy as well. This investigation revealed that the lncRNA MALAT1 (Metastasis-Associated Lung Adenocarcinoma Transcript 1), together with its enzymatic processing product called MALAT1-associated short cytoplasmic RNA, plays a significant role in providing favorable immunoregulatory capabilities in this context [62][63]. Increased levels of glucose are found to stimulate the expression of myocardial infarction-associated transcript (MIAT) and increase the expression of DAPK2 (Death-Associated Protein Kinase 2), promoting apoptosis in cardiomyocytes [64][65][66]. Furthermore, emerging evidence suggests that lncRNAs are involved in inflammation, oxidative stress, and angiogenesis, all of which play critical roles in HF pathophysiology [67]. Understanding the exact roles of these lncRNAs and their interactions with coding genes is important for understanding the molecular mechanisms of [68].
This entry is adapted from the peer-reviewed paper 10.3390/ncrna10010003
References
- Benjamin, E.J.; Muntner, P.; Alonso, A.; Bittencourt, M.S.; Callaway, C.W.; Carson, A.P.; Chamberlain, A.M.; Chang, A.R.; Cheng, S.; Das, S.R.; et al. Heart Disease and Stroke Statistics—2019 Update: A Report from the American Heart Association. Circulation 2019, 139, e56–e528.
- Ounzain, S.; Micheletti, R.; Beckmann, T.; Schroen, B.; Alexanian, M.; Pezzuto, I.; Crippa, S.; Nemir, M.; Sarre, A.; Johnson, R.; et al. Genome-Wide Profiling of the Cardiac Transcriptome after Myocardial Infarction Identifies Novel Heart-Specific Long Non-Coding RNAs. Eur. Heart J. 2015, 36, 353–368.
- Hangauer, M.J.; Vaughn, I.W.; McManus, M.T. Pervasive Transcription of the Human Genome Produces Thousands of Previously Unidentified Long Intergenic Noncoding RNAs. PLoS Genet. 2013, 9.
- Dhanoa, J.K.; Sethi, R.S.; Verma, R.; Arora, J.S.; Mukhopadhyay, C.S. Long Non-Coding RNA: Its Evolutionary Relics and Biological Implications in Mammals: A Review. J. Anim. Sci. Technol. 2018, 60, 25.
- Rinn, J.L.; Chang, H.Y. Genome Regulation by Long Noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166.
- Quinn, J.J.; Chang, H.Y. Unique Features of Long Non-Coding RNA Biogenesis and Function. Nat. Rev. Genet. 2016, 17, 47–62.
- Castel, S.E.; Martienssen, R.A. RNA Interference in the Nucleus: Roles for Small RNAs in Transcription, Epigenetics and Beyond. Nat. Rev. Genet. 2013, 14.
- Wu, T.; Du, Y. LncRNAs: From Basic Research to Medical Application. Int. J. Biol. Sci. 2017, 13, 295–307.
- Devaux, Y.; Zangrando, J.; Schroen, B.; Creemers, E.E.; Pedrazzini, T.; Chang, C.-P.; Dorn, G.W., II; Thum, T.; Heymans, S. Long Noncoding RNAs in Cardiac Development and Ageing. Nat. Rev. Cardiol. 2015, 12, 415–425.
- Bleumink, G.; Knetsch, A.; Sturkenboom, M.; Straus, S.; Hofman, A.; Deckers, J.; Witteman, J.; Stricker, B. Quantifying the Heart Failure Epidemic: Prevalence, Incidence Rate, Lifetime Risk and Prognosis of Heart Failure. Eur. Heart J. 2004, 25, 1614–1619.
- Paulus, W.J. Unfolding Discoveries in Heart Failure. N. Engl. J. Med. 2020, 382, 679–682.
- Packer, M. The Neurohormonal Hypothesis: A Theory to Explain the Mechanism of Disease Progression in Heart Failure. J. Am. Coll. Cardiol. 1992, 20, 248–254.
- McDonagh, T.A.; Metra, M.; Adamo, M.; Gardner, R.S.; Baumbach, A.; Böhm, M.; Burri, H.; Butler, J.; Čelutkienė, J.; Chioncel, O.; et al. 2021 ESC Guidelines for the Diagnosis and Treatment of Acute and Chronic Heart Failure. Eur. Heart J. 2021, 42, 3599–3726.
- Madhavan, M.; Gopalakrishnan, G. Long Non-Coding RNAs in Heart Failure: A Deep Belief Network Based Cluster Analysis. Curr. Bioinform. 2021, 16, 983–991.
- Fan, X.; Zhang, Z.; Zheng, L.; Wei, W.; Chen, Z. Long Non-Coding RNAs in the Pathogenesis of Heart Failure: A Literature Review. Front. Cardiovasc. Med. 2022, 9, 950284.
- Sallam, T.; Sandhu, J.; Tontonoz, P. Long Noncoding RNA Discovery in Cardiovascular Disease: Decoding Form to Function. Circ. Res. 2018, 122, 155–166.
- Greco, S.; Zaccagnini, G.; Perfetti, A.; Fuschi, P.; Valaperta, R.; Voellenkle, C.; Castelvecchio, S.; Gaetano, C.; Finato, N.; Beltrami, A.P.; et al. Long Noncoding RNA Dysregulation in Ischemic Heart Failure. J. Transl. Med. 2016, 14, 183.
- Han, Z.; Zhang, X.; Liu, C.; Lu, M.; Wang, J.; Nie, Y.; Zhang, H. Analysis of Long Noncoding RNAs Expression Profiles in the Human Cardiac Fibroblasts with Cardiac Fibrosis. Biochem. Biophys. Res. Commun. 2023, 660, 73–81.
- Lee, J.H.; Gao, C.; Peng, G.; Greer, C.; Ren, S.; Wang, Y.; Xiao, X. Analysis of Transcriptome Complexity through RNA Sequencing in Normal and Failing Murine Hearts. Circ. Res. 2011, 109, 1332–1341.
- Yang, K.C.; Yamada, K.A.; Patel, A.Y.; Topkara, V.K.; George, I.; Cheema, F.H.; Ewald, G.A.; Mann, D.L.; Nerbonne, J.M. Deep RNA Sequencing Reveals Dynamic Regulation of Myocardial Noncoding RNAs in Failing Human Heart and Remodeling with Mechanical Circulatory Support. Circulation 2014, 129, 1009–1021.
- Sheng, K.; Nie, Y.; Gao, B. Recent Advances in Myocardial Regeneration Strategy. J. Int. Med. Res. 2019, 47, 5453–5464.
- Ma, Y.; He, S.; Xie, Q.; Tang, Z.; Jiang, Z. LncRNA: An Important Regulator of Atherosclerosis. Curr. Med. Chem. 2023, 30, 4340–4354.
- Shen, L.; Shen, G.; Lu, X.; Ding, G.; Hu, X. Co-Expression Network Analysis Revealing the Potential Regulatory Roles of LncRNAs in Atrial Fibrillation. Curr. Bioinform. 2020, 15, 980–987.
- Li, M.; Zheng, H.; Han, Y.; Chen, Y.; Li, B.; Chen, G.; Chen, X.; Huang, S.; He, X.; Wei, G.; et al. LncRNA Snhg1-Driven Self-Reinforcing Regulatory Network Promoted Cardiac Regeneration and Repair after Myocardial Infarction. Theranostics 2021, 11, 9397–9414.
- Wang, H.; Lian, X.; Gao, W.; Gu, J.; Shi, H.; Ma, Y.; Li, Y.; Fan, Y.; Wang, Q.; Wang, L. Long Noncoding RNA H19 Suppresses Cardiac Hypertrophy through the MicroRNA-145-3p/SMAD4 Axis. Bioengineered 2022, 13, 3826–3839.
- Omura, J.; Habbout, K.; Shimauchi, T.; Wu, W.H.; Breuils-Bonnet, S.; Tremblay, E.; Martineau, S.; Nadeau, V.; Gagnon, K.; Mazoyer, F.; et al. Identification of Long Noncoding RNA H19 as a New Biomarker and Therapeutic Target in Right Ventricular Failure in Pulmonary Arterial Hypertension. Circulation 2020, 142, 1464–1484.
- Han, P.; Li, W.; Lin, C.-H.; Yang, J.; Shang, C.; Nurnberg, S.T.; Jin, K.K.; Xu, W.; Lin, C.-Y.; Lin, C.-J.; et al. A Long Noncoding RNA Protects the Heart from Pathological Hypertrophy. Nature 2014, 514, 102–106.
- Zhao, Z.; Li, X.; Gao, C.; Jian, D.; Hao, P.; Rao, L.; Li, M. Peripheral Blood Circular RNA Hsa_circ_0124644 Can Be Used as a Diagnostic Biomarker of Coronary Artery Disease. Sci. Rep. 2017, 7, 39918.
- Kumarswamy, R.; Bauters, C.; Volkmann, I.; Maury, F.; Fetisch, J.; Holzmann, A.; Lemesle, G.; de Groote, P.; Pinet, F.; Thum, T. Circulating Long Noncoding RNA, LIPCAR, Predicts Survival in Patients with Heart Failure. Circ. Res. 2014, 114, 1569–1575.
- Holdt, L.M.; Beutner, F.; Scholz, M.; Gielen, S.; Gäbel, G.; Bergert, H.; Schuler, G.; Thiery, J.; Teupser, D. ANRIL Expression Is Associated with Atherosclerosis Risk at Chromosome 9p21. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 620–627.
- Lo Sardo, V.; Chubukov, P.; Ferguson, W.; Kumar, A.; Teng, E.L.; Duran, M.; Zhang, L.; Cost, G.; Engler, A.J.; Urnov, F.; et al. Unveiling the Role of the Most Impactful Cardiovascular Risk Locus through Haplotype Editing. Cell 2018, 175, 1796–1810.
- Bergami, M.; Fabin, N.; Cenko, E.; Bugiardini, R.; Manfrini, O. MicroRNAs as Potential Biomarkers in Coronary Artery Disease. Curr. Top. Med. Chem. 2023, 23, 454–469.
- Bektik, E.; Cowan, D.B.; Wang, D.-Z. Long Non-Coding RNAs in Atrial Fibrillation: Pluripotent Stem Cell-Derived Cardiomyocytes as a Model System. Int. J. Mol. Sci. 2020, 21, 5424.
- Sangeeth, A.; Malleswarapu, M.; Mishra, A.; Gutti, R.K. Long Non-Coding RNA Therapeutics: Recent Advances and Challenges. Curr. Drug Targets 2022, 23, 1457–1464.
- Micheletti, R.; Plaisance, I.; Abraham, B.J.; Sarre, A.; Ting, C.-C.; Alexanian, M.; Maric, D.; Maison, D.; Nemir, M.; Young, R.A.; et al. The Long Noncoding RNA Wisper Controls Cardiac Fibrosis and Remodeling. Sci. Transl. Med. 2017, 9, eaai9118.
- Scheuermann, J.C.; Boyer, L.A. Getting to the Heart of the Matter: Long Non-Coding RNAs in Cardiac Development and Disease. EMBO J. 2013, 32, 1805–1816.
- Haemmig, S.; Feinberg, M.W. Targeting LncRNAs in Cardiovascular Disease. Circ. Res. 2017, 120, 620–623.
- Choong, O.K.; Lee, D.S.; Chen, C.-Y.; Hsieh, P.C.H. The Roles of Non-Coding RNAs in Cardiac Regenerative Medicine. Noncoding RNA Res. 2017, 2, 100–110.
- Zhang, Y.; Ye, F.; Xiong, D.; Gao, X. LDNFSGB: Prediction of Long Non-Coding Rna and Disease Association Using Network Feature Similarity and Gradient Boosting. BMC Bioinform. 2020, 21, 377.
- Yuan, T.; Krishnan, J. Non-Coding RNAs in Cardiac Regeneration. Front. Physiol. 2021, 12, 650566.
- Legnini, I.; Morlando, M.; Mangiavacchi, A.; Fatica, A.; Bozzoni, I. A Feedforward Regulatory Loop between HuR and the Long Noncoding RNA Linc-MD1 Controls Early Phases of Myogenesis. Mol. Cell 2014, 53, 506–514.
- Wang, Q.; Xie, Z. GAS5 Silencing Attenuates Hypoxia-induced Cardiomyocytes Injury by Targeting MiR-21/PTEN. Immun. Inflamm. Dis. 2023, 11, e945.
- Li, J.; Chen, Z.; Wang, X.; Song, H. LncRNA UCA1, MiR-26a, and MiR-195 in Coronary Heart Disease Patients: Correlation with Stenosis Degree, Cholesterol Levels, Inflammatory Cytokines, and Cell Adhesion Molecules. J. Clin. Lab. Anal. 2022, 36, e24070.
- Uchida, S.; Dimmeler, S. Long Noncoding RNAs in Cardiovascular Diseases. Circ. Res. 2015, 116, 737–750.
- Zhang, Y.; Jiao, L.; Sun, L.; Li, Y.; Gao, Y.; Xu, C.; Shao, Y.; Li, M.; Li, C.; Lu, Y.; et al. LncRNA ZFAS1 as a SERCA2a Inhibitor to Cause Intracellular Ca2+ Overload and Contractile Dysfunction in a Mouse Model of Myocardial Infarction. Circ. Res. 2018, 122, 1354–1368.
- Ishii, N.; Ozaki, K.; Sato, H.; Mizuno, H.; Saito, S.; Takahashi, A.; Miyamoto, Y.; Ikegawa, S.; Kamatani, N.; Hori, M.; et al. Identification of a Novel Non-Coding RNA, MIAT, That Confers Risk of Myocardial Infarction. J. Hum. Genet. 2006, 51, 1087–1099.
- Lopez-Crisosto, C.; Pennanen, C.; Vasquez-Trincado, C.; Morales, P.E.; Bravo-Sagua, R.; Quest, A.F.G.; Chiong, M.; Lavandero, S. Sarcoplasmic Reticulum–Mitochondria Communication in Cardiovascular Pathophysiology. Nat. Rev. Cardiol. 2017, 14, 342–360.
- Aung, L.H.H.; Chen, X.; Cueva Jumbo, J.C.; Li, Z.; Wang, S.; Zhao, C.; Liu, Z.; Wang, Y.; Li, P. Cardiomyocyte Mitochondrial Dynamic-Related LncRNA 1 (CMDL-1) May Serve as a Potential Therapeutic Target in Doxorubicin Cardiotoxicity. Mol. Ther. Nucleic Acids 2021, 25, 638–651.
- Cortassa, S.; Juhaszova, M.; Aon, M.A.; Zorov, D.B.; Sollott, S.J. Mitochondrial Ca2+, Redox Environment and ROS Emission in Heart Failure: Two Sides of the Same Coin? J. Mol. Cell. Cardiol. 2021, 151, 113–125.
- Sato, M.; Kadomatsu, T.; Miyata, K.; Warren, J.S.; Tian, Z.; Zhu, S.; Horiguchi, H.; Makaju, A.; Bakhtina, A.; Morinaga, J.; et al. The LncRNA Caren Antagonizes Heart Failure by Inactivating DNA Damage Response and Activating Mitochondrial Biogenesis. Nat. Commun. 2021, 12, 2529.
- Tian, J.; An, X.; Niu, L. Myocardial Fibrosis in Congenital and Pediatric Heart Disease. Exp. Ther Med. 2017, 13, 1660–1664.
- Lecerf, C.; Le Bourhis, X.; Adriaenssens, E. The Long Non-Coding RNA H19: An Active Player with Multiple Facets to Sustain the Hallmarks of Cancer. Cell. Mol. Life Sci. 2019, 76, 4673–4687.
- Yang, R.; Li, L.; Hou, Y.; Li, Y.; Zhang, J.; Yang, N.; Zhang, Y.; Ji, W.; Yu, T.; Lv, L.; et al. Long Non-Coding RNA KCND1 Protects Hearts from Hypertrophy by Targeting YBX1. Cell Death Dis. 2023, 14, 344.
- Wang, Z.; Zhang, X.-J.; Ji, Y.-X.; Zhang, P.; Deng, K.-Q.; Gong, J.; Ren, S.; Wang, X.; Chen, I.; Wang, H.; et al. The Long Noncoding RNA Chaer Defines an Epigenetic Checkpoint in Cardiac Hypertrophy. Nat. Med. 2016, 22, 1131–1139.
- Shimizu, I.; Minamino, T. Physiological and Pathological Cardiac Hypertrophy. J. Mol. Cell. Cardiol. 2016, 97, 245–262.
- Hinderer, S.; Schenke-Layland, K. Cardiac Fibrosis – A Short Review of Causes and Therapeutic Strategies. Adv. Drug Deliv. Rev. 2019, 146, 77–82.
- Hao, K.; Lei, W.; Wu, H.; Wu, J.; Yang, Z.; Yan, S.; Lu, X.-A.; Li, J.; Xia, X.; Han, X.; et al. LncRNA-Safe Contributes to Cardiac Fibrosis through Safe-Sfrp2-HuR Complex in Mouse Myocardial Infarction. Theranostics 2019, 9, 7282–7297.
- Bandesh, K.; Masih, D.; Bhattacharyya, N.; Bharadwaj, D. Harnessing the Potential of Long Non-Coding RNAs to Manage Metabolic Diseases. Curr. Pharm. Des. 2021, 27, 3668–3685.
- Liu, N.; Kataoka, M.; Wang, Y.; Pu, L.; Dong, X.; Fu, X.; Zhang, F.; Gao, F.; Liang, T.; Pei, J.; et al. LncRNA LncHrt Preserves Cardiac Metabolic Homeostasis and Heart Function by Modulating the LKB1-AMPK Signaling Pathway. Basic Res. Cardiol. 2021, 116, 48.
- Wu, H.; Liu, J.; Li, W.; Liu, G.; Li, Z. LncRNA-HOTAIR Promotes TNF-α Production in Cardiomyocytes of LPS-Induced Sepsis Mice by Activating NF-ΚB Pathway. Biochem. Biophys. Res. Commun. 2016, 471, 240–246.
- Gu, Q.; Wang, B.; Zhao, H.; Wang, W.; Wang, P.; Deng, Y. LncRNA Promoted Inflammatory Response in Ischemic Heart Failure through Regulation of MiR-455-3p/TRAF6 Axis. Inflamm. Res. 2020, 69, 667–681.
- Gast, M.; Rauch, B.H.; Nakagawa, S.; Haghikia, A.; Jasina, A.; Haas, J.; Nath, N.; Jensen, L.; Stroux, A.; Böhm, A.; et al. Immune System-Mediated Atherosclerosis Caused by Deficiency of Long Non-Coding RNA MALAT1 in ApoE−/− Mice. Cardiovasc. Res. 2019, 115, 302–314.
- Gast, M.; Schroen, B.; Voigt, A.; Haas, J.; Kuehl, U.; Lassner, D.; Skurk, C.; Escher, F.; Wang, X.; Kratzer, A.; et al. Long Noncoding RNA MALAT1-Derived MascRNA Is Involved in Cardiovascular Innate Immunity. J. Mol. Cell Biol. 2016, 8, 178–181.
- Sun, H.; Wang, C.; Zhou, Y.; Cheng, X. Long Noncoding RNA OIP5-AS1 Overexpression Promotes Viability and Inhibits High Glucose-Induced Oxidative Stress of Cardiomyocytes by Targeting MicroRNA-34a/SIRT1 Axis in Diabetic Cardiomyopathy. Endocr. Metab. Immune Disord. Drug Targets 2021, 21, 2017–2027.
- Zhou, X.; Zhang, W.; Jin, M.; Chen, J.; Xu, W.; Kong, X. LncRNA MIAT Functions as a Competing Endogenous RNA to Upregulate DAPK2 by Sponging MiR-22-3p in Diabetic Cardiomyopathy. Cell Death Dis. 2017, 8, e2929.
- Jia, G.; Hill, M.A.; Sowers, J.R. Diabetic Cardiomyopathy. Circ. Res. 2018, 122, 624–638.
- Kumar, M.M.; Goyal, R. LncRNA as a Therapeutic Target for Angiogenesis. Curr. Top. Med. Chem. 2017, 17, 1750–1757.
- Nair, N. Epidemiology and Pathogenesis of Heart Failure with Preserved Ejection Fraction. Rev. Cardiovasc. Med. 2020, 21, 531–540.
This entry is offline, you can click here to edit this entry!
