Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Viper venom phospholipase A2 enzymes (vvPLA2s) and phospholipase A2-like (PLA2-like) proteins are two of the principal toxins in viper venom that are responsible for the severe myotoxic and neurotoxic effects caused by snakebite envenoming, among other pathologies.
- snake venom viper
- phospholipase A2
- toxin
- myotoxicity
1. Epidemiology
The World Health Organization recognizes twenty neglected tropical diseases that affect the whole world, but their devastating effects are felt mostly in tropical areas and impoverished communities, where access to hospitals and medicines is limited [1][2]. These neglected tropical diseases can be divided into infectious diseases (19 diseases) and non-infectious diseases, of which there is only one—snakebite envenoming [3][4]. The mortality rate of neglected tropical diseases is ca. 200 thousand people per year, and they cause more than 19 million disabilities annually. In the last decade, the World Health Organization and governmental agencies have carried out several campaigns and investments, significantly reducing the number of infectious neglected tropical diseases. Nonetheless, the impact of snakebite envenoming remains alarming, with 81–138 thousand deaths yearly, the total of which may equal the number of deaths caused by the other 19 neglected tropical diseases altogether [5]. The intricate chemical and bioactive composition of snake venom means that it is challenging [6] to find a cure for snakebite envenoming. The current state-of-the-art treatment involves administering antibodies purified from the plasma of hyper-immunized equines [7][8]. However, it is very costly, and it requires transportation and storage in cold containers, as well as inpatient administration due to frequent anaphylactic reactions [9]. These constraints mean that this therapy is unavailable in the remote regions of resource-poor countries and in poor communities where most snakebites occur [1][7].
2. Venom Composition
Snake venoms are complex cocktails comprising tens to hundreds of components, of which >90% (w/w) are proteins and peptides [10][11][12]. The identity and abundance of components are diverse, and they depend on the snake species, gender, age, habitat, and prey, among other factors. Intraspecific variability is also significant. Venomous snakes belong to three families of front-fanged snakes, the Viperidae (vipers), Elapidae, and Atractaspidae, and a family of rear-fanged venomous snakes, the Colubridae; however, regarding the latter, not all members are venomous [13]. This work focuses primarily on viper venom, as vipers are estimated to be responsible for a substantial proportion of snakebite fatalities [14].
Despite the fact that viper venom is composed of highly diverse elements, three enzymatic toxin families stand out due to their toxicity and abundance: metalloproteinases (vvMPs), secreted phospholipase A2 enzymes (vvPLA2s), and serine proteases (vvSPs) [12]. These families frequently account for ca. 70% of the total venom weight. Other components that are often present, but in smaller amounts, are the C-type lectins, cysteine-rich secretory proteins, and L-amino acid oxidases [15] (for a more complete list, see Ref. [15]). vvMPs and vvPLA2s are present in more than 80% of the ca. 165 known viper venom proteomes [12].
3. Viper Venom Phospholipase A2 Enzymes
3.1. Pathophysiology of vvPLA2s
vvPLA2 is the second most abundant protein family in viper venom on average, surpassed only by vvMPs. It is thus evident that vvPLA2s and vvMPs are prime drug target candidates for treating snakebite envenoming. vvPLA2s play many pathophysiologic roles; regarding the ones found in viper venoms, the most common is myotoxicity [16][17][18][19][20]. Nevertheless, other usual effects caused by vvPLA2s include platelet aggregation, anticoagulation, hemolysis, cytotoxicity, and neurotoxicity [21][22]. They also participate in inflammatory processes and display bactericidal activities [23]. As a whole, viper envenomation produces highly harmful consequences, such as muscle breakdown, neuromuscular paralysis, tissue necrosis and amputations, bleeding disorders, thrombosis, kidney and heart failure, hypovolemic shock, and often death [24][25][26][27], effects for which vvPLA2 is at least partially responsible.
3.2. The Structure of vvPLA2
vvPLA2s are small, secreted proteins of 13–15 kDa. Together with the human synovial fluid PLA2, they form the PLA2 group IIA [28][29][30]. Within this group, almost all enzymes have seven disulfide bonds, significant sequence identity, and a common fold (Figure 1). Rare exceptions within the viper family include the Gaboon viper (Bitis gabonica), the Rhinoceros viper (Bitis nasicornis) and Palm pit vipers (Trimeresurus genus) vvPLA2s.
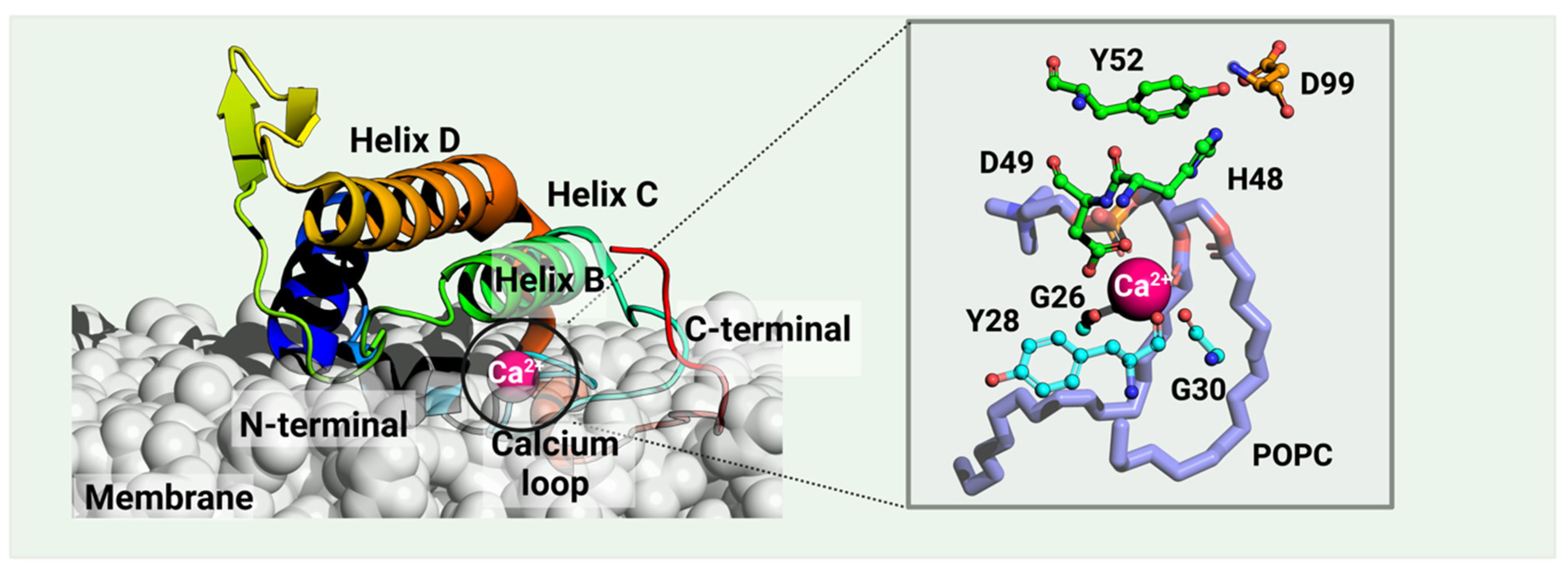
Figure 1. A catalytic vvPLA2 (Echis carinatus, PDB ID: 1OZ6 [31], in rainbow) bound on a phospholipid bilayer cell membrane (white) is shown. The Methods section provides the details of the protein–membrane complex modeling. The phospholipid bound to the catalytic cavity is represented by lilac sticks. Residues H48 and D99 compose the catalytic dyad, the latter stabilized by Y52. vvPLA2s need a Ca2+ cofactor for catalytic activity, which is heptacoordinated by D49 (double coordination), three peptide carbonyl oxygens of the “calcium loop” (residues G26, Y28, and G30), and the substrate’s phosphate and sn-2 ester bond. Residue numbering follows the standard numbering of Renetseder et al. [32].
The vvPLA2 enzymes are further characterized by their isoelectric point. Accordingly, acidic vvPLA2s typically have an isoelectric point of 4.0–5.5 with many acidic, negative residues, and basic vvPLA2s have an isoelectric point of ca. 8.0 with primarily basic and positive protein surface residues. The isoelectric point influences the vvPLA2 toxicity, and as a consequence, basic enzymes are far more toxic than acidic ones. The underlying reason still needs to be clarified but is supposed to lie in the different affinities that the acidic and basic enzymes have for the cell membrane phospholipids and/or for the cell membrane protein receptors.
3.3. The Catalytic Activity of vvPLA2
vvPLA2 enzymes cleave phospholipids at position sn-2, releasing fatty acids and lysophosphatidic acids. The corresponding mechanism of this hydrolysis reaction is only partially understood at the atomic level, occurring always in the presence of a Ca2+ cofactor. The latter is coordinated to Asp49 (bidentate), three carbonyl groups belonging to both a glycine and a tyrosine or phenylalanine residues located in the Ca2+-binding loop, and the substrate’s phosphate and sn-2 carbonyl groups (or two water molecules when in the unbound state). According to the postulated mechanisms, His48 deprotonates one water molecule, either directly (“single-water mechanism” hypothesis) or through a second bridging water molecule (“assisted water mechanism” hypothesis), generating a hydroxide ion that attacks the sn-2 carbon of the phospholipid (Figure 2A). The hydrogen bonding of His48 to the carboxylate of Asp99 stabilizes the positive form of His48, facilitating water deprotonation. Whether or not the nucleophilic water molecule is Ca2+-bound is unclear. Anyhow, the Ca2+ stabilizes the reaction’s transition state by coordinating the oxyanion generated at the sn-2 carbonyl site (and eventually the hydroxide nucleophile), subsequently decaying into a tetrahedral intermediate (Figure 2B). Following the ester bond cleavage, the hydrolyzed products—fatty acid and lysophospholipid—are released (Figure 2C). No evidence exists for protein conformational changes during the cycle [33].
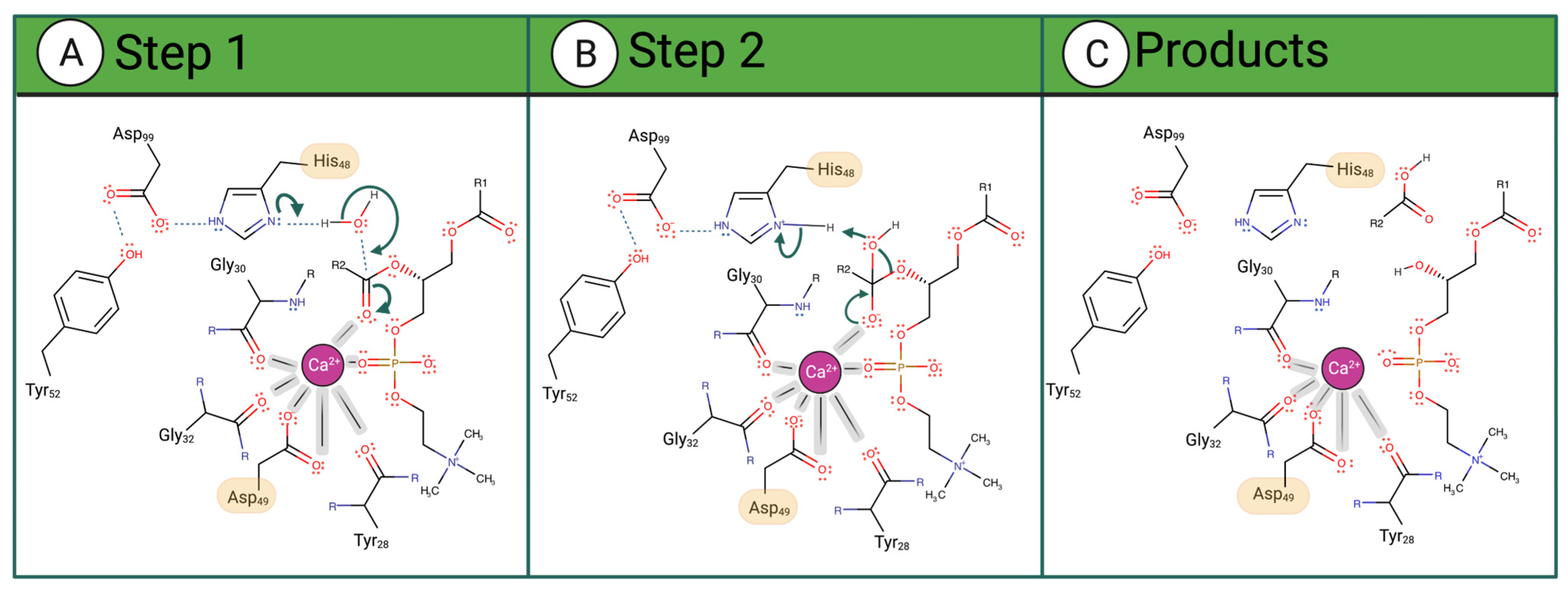
Figure 2. Proposed catalytic single-water mechanism of sPLA2 for a phosphatidylcholine substrate. The reaction occurs in 3 steps: (A) His48 deprotonates a water molecule whose hydroxide ion attacks the substrate sn-2 carbon (B) The oxyanion collapses, eliminating the phosphocholine substrate that deprotonates His48. (C) The fatty acid and the lysophosphatidylcholine are released as products.
3.4. The vvPLA2 Protein–Membrane Interface
Monomeric and dimeric vvPLA2s are the most common quaternary structures found via X-ray crystallography [19][34][35][36], and they are supposed to be the most common in solution too.
Two different dimeric quaternary structures have been found in cristallo, named “conventional” and “alternative” [37]. Nevertheless, the researchers will refer to them here as “extended” and “compact” dimers, respectively, as this nomenclature better reflects the nature of their quaternary structure, and the fact that the buried area of these two structures is very different. The extended dimer of a PLA2-like protein (i.e., a non-enzymatic homolog of the vvPLA2 enzymes, as discussed in Section 3.6), from, e.g., B. pauloensis, buries an area of 1022 Å2 (Ref. [38]), and it has few intermonomer contacts, which may turn the dimerization in water less robust. Nevertheless, it has phospholipid binding sites open to the solvent. The compact conformation of the same enzyme achieves a larger buried area of 1491 Å2 (Ref. [39]) and establishes many more intermonomer contacts than the extended conformation. It seems more stable in water, even though each monomer occludes the binding site of the other. Thus, a conformational change or dissociation may be needed to expose the binding site of a compact dimer to the cell membrane.
vvPLA2s and similar human pancreatic PLA2s are believed to bind the cell membrane as monomers in most cases [40]. Therefore, if a PLA2 forms dimers in solution, dimer dissociation should precede or happen concomitantly with membrane binding. vvPLA2s have more affinity for membrane regions richer in negative than zwitterionic phospholipids, particularly the most toxic basic isoforms. Insights from an anion-assisted dimer of pancreatic porcine PLA2 [41][42] estimate that PLA2 buries 30–40 phospholipids, which form salt bridges with the PLA2 basic residues [43]. Acidic vvPLA2s are generally catalytically more active in vitro but less toxic in vivo than the basic isoforms [44]. In contrast, most basic vvPLA2s induce several toxic effects [17][26][37][45][46].
3.5. The Role of the N-Terminal Region for Membrane Binding and Enzymatic Activity
vvPLA2s contain an α-helical N-terminal region believed to be implicated in critical functions, such as membrane binding, and vital structural areas, such as the substrate-binding hydrophobic channel. X-ray structures of PLA2s from the Indian cobra (Naja naja) [47] and the European bee (Apis mellifera) [48] venoms and from human [49] and pig (sus scrofa) [50] secretory PLA2s, solved in the presence of transition-state analogs, showed the N-terminal region interacting with the phospholipid substrate. The invariant residues Leu2, Phe5, and Ile9 delimitate the substrate-binding cavity on one side. The contribution of the N-terminal region to the catalytic activity has been extensively studied in mammalian PLA2s, where the deletion of 8–10 residues of the N-terminus resulted in a nearly complete loss of enzyme activity [51][52]. Moreover, deleting the ten N-terminal residues made the enzyme bind to membranes with lower affinity and random, non-specific orientations [53]. Such studies underline the importance of the N-terminal region in regulating the PLA2 function by determining the strength of membrane binding and the productive orientation of PLA2 at the membrane surface and in contributing to possible structural changes in the enzyme during interfacial activation [54][55]. However, few studies regarding this topic were performed with vvPLA2. Nevertheless, studies involving a vvPLA2 from Crotalus atrox confirmed that deleting the ten N-terminal residues results in a loss of enzymatic activity as well as the dimeric structure of the native enzyme [56].
3.6. The PLA2-Like Proteins and Their Myotoxic C-Terminal Region
There are two types of PLA2 proteins in viper venom: the previously discussed Ca2+-dependent catalytically active enzymes (vvPLA2) and the non-enzymatic PLA2 proteins (Figure 3). The latter are named PLA2-homologs, Lys-49 PLA2, or PLA2-like proteins (the last designation will be used here) and are exclusively found in viper venoms as they have diverged from the former during evolution.
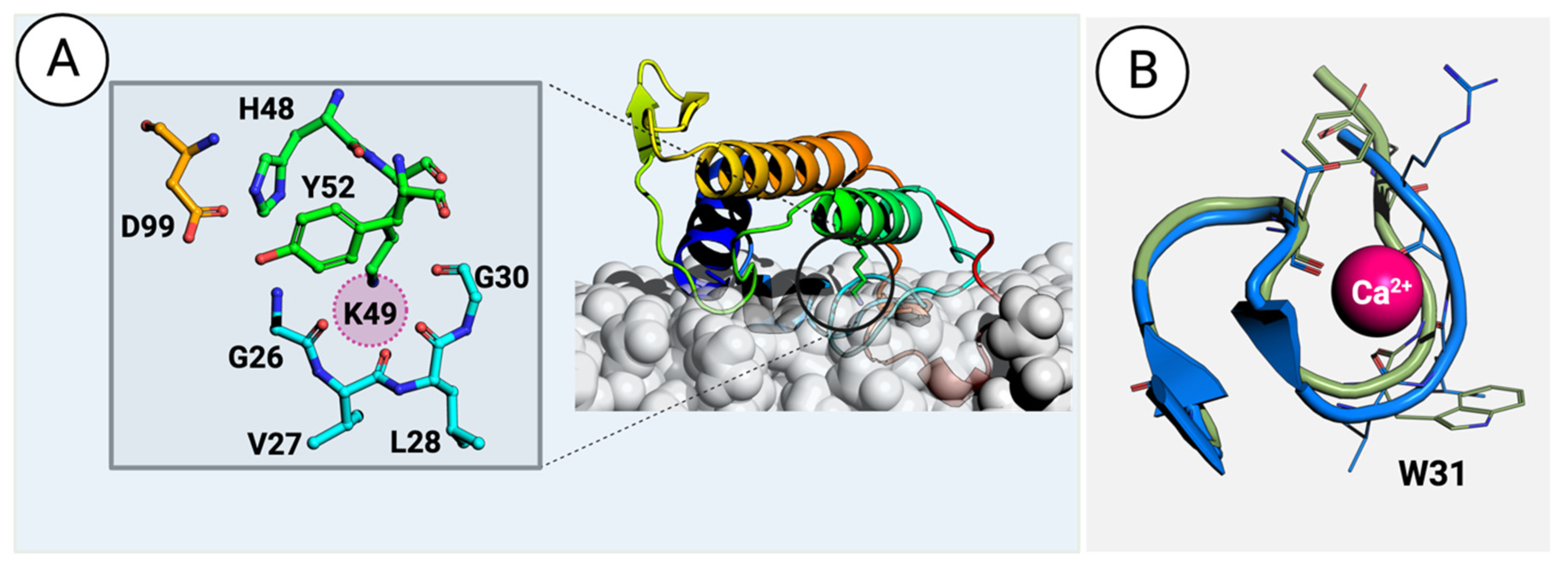
Figure 3. (A) A PLA2-like protein from the venom of Bothrops asper (PDB ID: 1Y4L [57]) interacts with a phospholipid bilayer membrane. The details of modeling the protein–membrane complex are given in the Methods section. The inset on the left shows the residues surrounding and stabilizing the H48K49C50C51 motif. The sphere in magenta highlights the position occupied by the calcium atom in vvPLA2. (B) Superimposition of the vvPLA2 Ca2+ loop (green) on the equivalent loop region in the PLA2-like proteins (blue). The small difference in the position of the loop is sufficient to affect the binding of the Ca2+ ion.
The main differences found between vvPLA2 and PLA2-like proteins are as follows:
- i.
-
The residue at position 49, which can be Asp in the enzymes and Lys or, more rarely, Ser, Asn, Gln, or Arg, in the PLA2-like proteins);
- ii.
-
The active site Ca2+ cofactor, which is only present in vvPLA2;
- iii.
-
The sequence and fold of the Ca2+-binding loop;
- iv.
PLA2-like proteins have been extensively studied for their ability to disrupt the cell membrane, causing myotoxicity [16][58][59][60][61][62][63][64]. A large body of studies recently reviewed in Ref. [57] identifies the C-terminal region (residues 105–117) in the mature Bothrops asper myotoxin II, Uniprot ID: P24605, or equivalently residues 115–129 when using the standard numbering (i.e., a common residue numbering for mammalian pancreatic and venom PLA2 enzymes [32]), composed primarily of cationic, hydrophobic, and aromatic residues, as the primary determinant for myotoxicity. Moreover, the same region is involved in hyperalgesia and inflammation. In several instances, peptides with the 115–129 sequence retain, partly or wholly, the bioactivity of the complete enzyme [60]. However, given the differences between the bioactivity of the total enzyme and the 115–129 peptide, studies suggest that the C-terminal region is central but not solely responsible for PLA2 toxicity, with other residues, such as K20, K36, and K38, also being crucial for the effect [16][45][64][65]. Like vvPLA2, PLA2-like proteins induce local membrane perturbations, allowing Ca2+, K+, and ATP to be internalized through membrane crossing, leading to extreme cytotoxic events [66][67].
3.7. Evaluation of vvPLA2 Druggability
Researchers are developing small-molecule drug treatments against snakebite envenoming to overcome the limitations of the antibody-based treatment [68][69][70]. The vvPLA2s are a primary drug target because they are expressed in more than 90% of viper species [10][11][12] and are associated with drastic pathologic processes [21][71][72]. Moreover, it is known that competitive enzymatic inhibitors such as varespladib [69][73][74][75][76][77][78][79] also inhibit PLA2-like myotoxicity [69][74][80].
This entry is adapted from the peer-reviewed paper 10.3390/toxins16020071
References
- Harrison, R.A.; Hargreaves, A.; Wagstaff, S.C.; Faragher, B.; Lalloo, D.G. Snake Envenoming: A Disease of Poverty. PLoS Negl. Trop. Dis. 2009, 3, e569.
- The Lancet. Snake-bite envenoming: A priority neglected tropical disease. Lancet 2017, 390, 2.
- WHO. The Selection and Use of Essential Medicines: Report of the WHO Expert Committee on Selection and Use of Essential Medicines; Including the 22nd WHO Model List of Essential Medicines and the 8th WHO Model List of Essential Medicines for Children; WHO Technical Report Series, No. 1035; World Health Organization: Geneva, Switzerland, 2021.
- WHO. Snakebite Envenoming: A Strategy for Prevention and Control; World Health Organization: Geneva, Switzerland, 2019.
- Gutiérrez, J.M.; Calvete, J.J.; Habib, A.G.; Harrison, R.A.; Williams, D.J. Snakebite envenoming. Nat. Rev. Dis. Primers 2017, 3, 17063.
- Casewell, N.R.; Wüster, W.; Vonk, F.J.; Harrison, R.A.; Fry, B.G. Complex cocktails: The evolutionary novelty of venoms. Trends Ecol. Evol. 2013, 28, 219–229.
- Longbottom, J.; Shearer, F.M.; Devine, M.; Alcoba, G.; Chappuis, F.; Weiss, D.J.; Ray, S.E.; Ray, N.; Warrell, D.A.; de Castañeda, R.R.; et al. Vulnerability to snakebite envenoming: A global mapping of hotspots. Lancet 2018, 392, 673.
- Alirol, E.; Lechevalier, P.; Zamatto, F.; Chappuis, F.; Alcoba, G.; Potet, J. Antivenoms for Snakebite Envenoming: What Is in the Research Pipeline? PLoS Negl. Trop. Dis. 2015, 9, e0003896.
- Puzari, U.; Mukherjee, A.K. Recent developments in diagnostic tools and bioanalytical methods for analysis of snake venom: A critical review. Anal. Chim. Acta 2020, 1137, 208–224.
- Tasoulis, T.; Isbister, G.K. A review and database of snake venom proteomes. Toxins 2017, 9, 290.
- Tasoulis, T.; Pukala, T.L.; Isbister, G.K. Investigating Toxin Diversity and Abundance in Snake Venom Proteomes. Front. Pharmacol. 2022, 12, 768015.
- Oliveira, A.L.; Viegas, M.F.; da Silva, S.L.; Soares, A.M.; Ramos, M.J. The chemistry of snake venom and its medicinal potential. Nat. Rev. Chem. 2022, 6, 451–469.
- Ossiboff, R.J. Serpentes. In Pathology of Wildlife and Zoo Animals; Academic Press: Cambridge, MA, USA, 2018; pp. 897–919.
- Gopalakrishnan, M.; Saurabh, S.; Sagar, P.; Bammigatti, C.; Dutta, T.K. A simple mortality risk prediction score for viper envenoming in India (VENOMS): A model development and validation study. PLoS Negl. Trop. Dis. 2022, 16, e0010183.
- Calvete, J.J.; Lomonte, B.; Saviola, A.J.; Bonilla, F.; Sasa, M.; Williams, D.J.; Undheim, E.A.B.; Sunagar, K.; Jackson, T.N.W. Mutual enlightenment: A toolbox of concepts and methods for integrating evolutionary and clinical toxinology via snake venomics and the contextual stance. Toxicon X 2021, 10, 100070.
- Lomonte, B.; Angulo, Y.; Calderón, L. An overview of lysine-49 phospholipase A2 myotoxins from crotalid snake venoms and their structural determinants of myotoxic action. Toxicon 2003, 42, 885–901.
- Gutiérrez, J.M.; Ownby, C.L. Skeletal muscle degeneration induced by venom phospholipases A2: Insights into the mechanisms of local and systemic myotoxicity. Toxicon 2003, 42, 915–931.
- Ohno, M.; Chijiwa, T.; Oda-Ueda, N.; Ogawa, T.; Hattori, S. Molecular evolution of myotoxic phospholipases A2 from snake venom. Toxicon 2003, 42, 841–854.
- Ullah, A.; Souza TAC, B.; Betzel, C.; Murakami, M.T.; Arni, R.K. Crystallographic portrayal of different conformational states of a Lys49 phospholipase A2 homologue: Insights into structural determinants for myotoxicity and dimeric configuration. Int. J. Biol. Macromol. 2012, 51, 209–214.
- Angulo, Y.; Lomonte, B. Inhibitory effect of fucoidan on the activities of crotaline snake venom myotoxic phospholipases A2. Biochem. Pharmacol. 2003, 66, 1993–2000.
- Gutiérrez, J.M.; Lomonte, B. Phospholipases A(2): Unveiling the secrets of a functionally versatile group of snake venom toxins. Toxicon 2013, 62, 27–39.
- Xiao, H.; Pan, H.; Liao, K.; Yang, M.; Huang, C. Snake Venom PLA2, a Promising Target for Broad-Spectrum Antivenom Drug Development. Biomed. Res. Int. 2017, 2017, 6592820.
- Costa, T.R.; Menaldo, D.L.; Oliveira, C.Z.; Santos-Filho, N.A.; Teixeira, S.S.; Nomizo, A.; Fuly, A.L.; Monteiro, M.C.; de Souza, B.M.; Palma, M.S.; et al. Myotoxic phospholipases A(2) isolated from Bothrops brazili snake venom and synthetic peptides derived from their C-terminal region: Cytotoxic effect on microorganism and tumor cells. Peptides 2008, 29, 1645–1656.
- Harris, J.B.; Scott-Davey, T. Secreted phospholipases A2 of snake venoms: Effects on the peripheral neuromuscular system with comments on the role of phospholipases A2 in disorders of the CNS and their uses in industry. Toxins 2013, 5, 2533–2571.
- Borges, R.J.; Lemke, N.; Fontes, M.R.M. PLA2-like proteins myotoxic mechanism: A dynamic model description. Sci. Rep. 2017, 7, 15514.
- Salvador, G.H.M.; dos Santos, J.I.; Lomonte, B.; Fontes, M.R.M. Crystal structure of a phospholipase A2 from Bothrops asper venom: Insights into a new putative “myotoxic cluster”. Biochimie 2017, 133, 95–102.
- Fernandes, C.A.H.; Comparetti, E.J.; Borges, R.J.; Huancahuire-Vega, S.; Ponce-Soto, L.A.; Marangoni, S.; Soares, A.M.; Fontes, M.R. Structural bases for a complete myotoxic mechanism: Crystal structures of two non-catalytic phospholipases A2-like from Bothrops brazili venom. Biochim. Biophys. Acta Proteins Proteom. 2013, 1834, 2772–2781.
- Six, D.A.; Dennis, E.A. The expanding superfamily of phospholipase A(2) enzymes: Classification and characterization. Biochim. Biophys. Acta 2000, 1488, 1–19.
- Schaloske, R.H.; Dennis, E.A. The phospholipase A2 superfamily and its group numbering system. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2006, 1761, 1246–1259.
- Burke, J.E.; Dennis, E.A. Phospholipase A2 biochemistry. Cardiovasc. Drugs Ther. 2009, 23, 49–59.
- Jasti, J.; Paramasivam, M.; Srinivasan, A.; Singh, T.P. Structure of an acidic phospholipase A2 from Indian saw-scaled viper (Echis carinatus) at 2.6 Å resolution reveals a novel intermolecular interaction. Acta Crystallogr. D Biol. Crystallogr. 2003, 60, 66–72.
- Renetseder, R.; Brunie, S.; Dijkstra, B.W. A comparison of the crystal structure of phospholipase A2 from bovine pancreas and Crotalus atrox venom. J. Biol. Chem. 1985, 260, 11627–11634.
- Castro-Amorim, J.; de Oliveira, A.N.; Da Silva, S.L.; Soares, A.M.; Mukherjee, A.K.; Ramos, M.J.; Fernandes, P.A. Catalytically Active Snake Venom PLA2 Enzymes: An Overview of Its Elusive Mechanisms of Reaction. J. Med. Chem. 2023, 66, 5364–5376.
- Matsui, T.; Kamata, S.; Ishii, K.; Maruno, T.; Ghanem, N.; Uchiyama, S.; Kato, K.; Suzuki, A.; Oda-Ueda, N.; Ogawa, T.; et al. SDS-induced oligomerization of Lys49-phospholipase A2 from snake venom. Sci. Rep. 2019, 9, 2330.
- Gomes, A.A.S.; Cardoso, F.F.; Souza, M.F.; Oliveira, C.L.P.; Perahia, D.; Magro, A.J.; Fontes, M.R.M. The allosteric activation mechanism of a phospholipase A2-like toxin from Bothrops jararacussu venom: A dynamic description. Sci. Rep. 2020, 10, 16252.
- Borges, R.J.; Cardoso, F.F.; de Carvalho, C.; de Marino, I.; Pereira, P.S.; Soares, A.M.; Dal-Pai-Silva, M.; Usón, I.; Fontes, M.R. Structural and functional studies of a snake venom phospholipase A2-like protein complexed to an inhibitor from Tabernaemontana catharinensis. Biochimie 2023, 206, 105–115.
- dos Santos, J.I.; Soares, A.M.; Fontes, M.R.M. Comparative structural studies on Lys49-phospholipases A2 from Bothrops genus reveal their myotoxic site. J. Struct. Biol. 2009, 167, 106–116.
- Magro, A.J.; Soares, A.M.; Giglio, J.R.; Fontes, M.R.M. Crystal structures of BnSP-7 and BnSP-6, two Lys49-phospholipases A2: Quaternary structure and inhibition mechanism insights. Biochem. Biophys. Res. Commun. 2003, 311, 713–720.
- de Lima, L.F.G.; Borges, R.J.; Viviescas, M.A.; Fernandes, C.A.H.; Fontes, M.R.M. Structural studies with BnSP-7 reveal an atypical oligomeric conformation compared to phospholipases A2-like toxins. Biochimie 2017, 142, 11–21.
- Jain, M.K.; Rogers, J. Substrate specificity for interfacial catalysis by phospholipase A2 in the scooting mode. Biochim. Et Biophys. Acta (BBA)/Lipids Lipid Metab. 1989, 1003, 91–97.
- Pan, Y.H.; Epstein, T.M.; Jain, M.K.; Bahnson, B.J. Five coplanar anion binding sites on one face of phospholipase A2: Relationship to interface binding. Biochemistry 2001, 40, 609–617.
- Pan, Y.H.; Yu, B.Z.; Berg, O.G.; Jain, M.K.; Bahnson, B.J. Crystal structure of phospholipase A2 complex with the hydrolysis products of platelet activating factor: Equilibrium binding of fatty acid and lysophospholipid-ether at the active site may be mutually exclusive. Biochemistry 2002, 41, 14790–14800.
- Bahnson, B.J. Structure, function and interfacial allosterism in phospholipase A2: Insight from the anion-assisted dimer. Arch. Biochem. Biophys. 2005, 433, 96–106.
- Fernández, J.; Gutiérrez, J.M.; Angulo, Y.; Sanz, L.; Juárez, P.; Calvete, J.J.; Lomonte, B. Isolation of an acidic phospholipase A2 from the venom of the snake Bothrops asper of Costa Rica: Biochemical and toxicological characterization. Biochimie 2010, 92, 273–283.
- Fernandes, C.A.H.; Borges, R.J.; Lomonte, B.; Fontes, M.R.M. A structure-based proposal for a comprehensive myotoxic mechanism of phospholipase A2-like proteins from viperid snake venoms. Biochim. Et Biophys. Acta (BBA) Proteins Proteom. 2014, 1844, 2265–2276.
- Lomonte, B.; Gutiérrez, J.M. Phospholipases A2 From Viperidae Snake Venoms: How do They Induce Skeletal Muscle Damage? Acta Chim. Slov. 2011, 58, 647–659.
- White, S.P.; Scott, D.L.; Otwinowski, Z.; Gelb, M.H.; Sigler, P.B. Crystal structure of cobra-venom phospholipase A2 in a complex with a transition-state analogue. Science 1990, 250, 1560–1563.
- Scott, D.L.; Otwinowski, Z.; Gelb, M.H.; Sigler, P.B. Crystal structure of bee-venom phospholipase A2 in a complex with a transition-state analogue. Science 1990, 250, 1563–1566.
- Scott, D.L.; White, S.P.; Browning, J.L.; Rosa, J.J.; Gelb, M.H.; Sigler, P.B. Structures of Free and Inhibited Human Secretory Phospholipase A2 from Inflammatory Exudate. Science 1991, 254, 1007–1010.
- Thunnissen, M.M.G.M.; AB, E.; Kalk, K.H.; Drenth, J.; Dijkstra, B.W.; Kuipers, O.P.; Dijkman, R.; de Haas, G.H.; Verheij, H.M. X-ray structure of phospholipase A2 complexed with a substrate-derived inhibitor. Nature 1990, 347, 689–691.
- Dijkstra, B.W.; Kalk, K.H.; Drenth, J.; De Hass, G.H.; Egmond, M.R.; Slotboom, A.J. Role of the N-Terminus in the Interaction of Pancreatic Phospholipase A2 with Aggregated Substrates. Properties and Crystal Structure of Transaminated Phospholipase A2. Biochemistry 1984, 23, 2759–2766.
- Liu, X.; Zhu, H.; Huang, B.; Rogers, J.; Yu, B.-Z.; Kumar, A.; Jain, M.K.; Sundaralingam, M.; Tsai, M.-D. Phospholipase A2 engineering. Probing the structural and functional roles of N-terminal residues with site-directed mutagenesis, X-ray, and NMR. Biochemistry 1995, 34, 7322–7334.
- Maliwal, B.P.; Yu, B.Z.; Szmacinski, H.; Squier, T.; van Binsbergen, J.; Slotboom, A.J.; Jain, M.K. Functional Significance of the Conformational Dynamics of the N-Terminal Segment of Secreted Phospholipase A2 at the Interface. Biochemistry 1994, 33, 4509–4516.
- Van Wezel, F.M.; Slotboom, A.J.; De Haas, G.H. Studies on the role of methionine in porcine pancreatic phospholipase A2. Biochim. Et Biophys. Acta 1976, 452, 101–111.
- Chiou, Y.L.; Cheng, Y.C.; Kao, P.H.; Wang, J.J.; Chang, L.-S. Mutations on the N-terminal region abolish differentially the enzymatic activity, membrane-damaging activity and cytotoxicity of Taiwan cobra phospholipase A2. Toxicon 2008, 51, 270–279.
- Randolph, A.; Heinrikson, R.L. Crotalus atrox phospholipase az amino acid sequence and studies on the function of the NH2-terminal region. J. Biol. Chem. 1982, 257, 2155–2161.
- Murakami, M.T.; Arruda, E.Z.; Melo, P.A.; Martinez, A.B.; Calil-Eliás, S.; Tomaz, M.A.; Lomonte, B.; Gutiérrez, J.M.; Arni, R.K. Inhibition of myotoxic activity of Bothrops asper myotoxin II by the anti-trypanosomal drug suramin. J. Mol. Biol. 2005, 350, 416–426.
- Mora-Obando, D.; Fernández, J.; Montecucco, C.; Gutiérrez, J.M.; Lomonte, B. Synergism between basic Asp49 and Lys49 phospholipase A2 myotoxins of viperid snake venom in vitro and in vivo. PLoS ONE 2014, 9, e109846.
- Lomonte, B. Lys49 myotoxins: Emerging insights into their modes of action. Toxicon 2020, 177, S5.
- Lomonte, B. Lys49 myotoxins, secreted phospholipase A2-like proteins of viperid venoms: A comprehensive review. Toxicon 2023, 224, 107024.
- Lomonte, B.; Rangel, J. Snake venom Lys49 myotoxins: From phospholipases A2 to non-enzymatic membrane disruptors. Toxicon 2012, 60, 520–530.
- Fernández, J.; Caccin, P.; Koster, G.; Lomonte, B.; Gutiérrez, J.M.; Montecucco, C.; Postle, A.D. Muscle phospholipid hydrolysis by Bothrops asper Asp49 and Lys49 phospholipase A2 myotoxins—Distinct mechanisms of action. FEBS J. 2013, 280, 3878–3886.
- Núez, C.E.; Angulo, Y.; Lomonte, B. Identification of the myotoxic site of the Lys49 phospholipase A2 from Agkistrodon piscivorus piscivorus snake venom: Synthetic C-terminal peptides from Lys49, but not from Asp49 myotoxins, exert membrane-damaging activities. Toxicon 2001, 39, 1587–1594.
- Zambelli, V.O.; Chioato, L.; Gutierrez, V.P.; Ward, R.J.; Cury, Y. Structural determinants of the hyperalgesic activity of myotoxic Lys49-phospholipase A2. J. Venom. Anim. Toxins Incl. Trop. Dis. 2017, 23, 7.
- Chioato, L.; Aragão, E.A.; Ferreira, T.L.; de Medeiros, A.I.; Faccioli, L.H.; Ward, R.J. Mapping of the structural determinants of artificial and biological membrane damaging activities of a Lys49 phospholipase A2 by scanning alanine mutagenesis. Biochim. Biophys. Acta Biomembr. 2007, 1768, 1247–1257.
- Cintra-Francischinelli, M.; Caccin, P.; Chiavegato, A.; Pizzo, P.; Carmignoto, G.; Angulo, Y.; Lomonte, B.; Gutiérrez, J.M.; Montecucco, C. Bothrops snake myotoxins induce a large efflux of ATP and potassium with spreading of cell damage and pain. Proc. Natl. Acad. Sci. USA 2010, 107, 14140–14145.
- Ivanušec, A.; Šribar, J.; Križaj, I. Secreted Phospholipases A2—Not just Enzymes: Revisited. Int. J. Biol. Sci. 2022, 18, 873–888.
- Bulfone, T.C.; Samuel, S.P.; Bickler, P.E.; Lewin, M.R. Developing Small Molecule Therapeutics for the Initial and Adjunctive Treatment of Snakebite. J. Trop. Med. 2018, 2018, 4320175.
- Salvador, G.H.M.; Borges, R.J.; Lomonte, B.; Lewin, M.R.; Fontes, M.R.M. The synthetic varespladib molecule is a multi-functional inhibitor for PLA2 and PLA2-like ophidic toxins. Biochim. Biophys. Acta Gen. Subj. 2021, 1865, 129913.
- Puzari, U.; Fernandes, P.A.; Mukherjee, A.K. Advances in the Therapeutic Application of Small-Molecule Inhibitors and Repurposed Drugs against Snakebite. J. Med. Chem. 2021, 64, 13938–13979.
- Kini, R.M.; Evans, H.J. A model to explain the pharmacological effects of snake venom phospholipases A2. Toxicon 1989, 27, 613–635.
- Kini, R.M. Excitement ahead: Structure, function and mechanism of snake venom phospholipase A2 enzymes. Toxicon 2003, 42, 827–840.
- Gutiérrez, J.M.; Lewin, M.R.; Williams, D.J.; Lomonte, B. Varespladib (LY315920) and methyl varespladib (LY333013) abrogate or delay lethality induced by presynaptically acting neurotoxic snake venoms. Toxins 2020, 12, 131.
- Salvador, G.H.M.; Gomes, A.A.S.; Bryan-Quirós, W.; Fernández, J.; Lewin, M.R.; Gutiérrez, J.M.; Lomonte, B.; Fontes, M.R.M. Structural basis for phospholipase A2-like toxin inhibition by the synthetic compound Varespladib (LY315920). Sci. Rep. 2019, 9, 17203.
- Zinenko, O.; Tovstukha, I.; Korniyenko, Y. PLA2 inhibitor varespladib as an alternative to the antivenom treatment for bites from nikolsky’s viper Vipera berus nikolskii. Toxins 2020, 12, 356.
- Lewin, M.; Samuel, S.; Merkel, J.; Bickler, P. Varespladib (LY315920) Appears to Be a Potent, Broad-Spectrum, Inhibitor of Snake Venom Phospholipase A2 and a Possible Pre-Referral Treatment for Envenomation. Toxins 2016, 8, 248.
- Lay, M.; Liang, Q.; Isbister, G.K.; Hodgson, W.C. In Vitro Efficacy of Antivenom and Varespladib in Neutralising Chinese Russell’s Viper (Daboia siamensis) Venom Toxicity. Toxins 2023, 15, 62.
- Villalta, M.; Pla, D.; Yang, S.L.; Sanz, L.; Segura, A.; Vargas, M.; Chen, P.Y.; Herrera, M.; Estrada, R.; Cheng, Y.F.; et al. Snake venomics and antivenomics of Protobothrops mucrosquamatus and Viridovipera stejnegeri from Taiwan: Keys to understand the variable immune response in horses. J. Proteom. 2012, 75, 5628–5645.
- Gutierres, P.G.; Pereira, D.R.; Vieira, N.L.; Arantes, L.F.; Silva, N.J.; Torres-Bonilla, K.A.; Hyslop, S.; Morais-Zani, K.; Nogueira, R.M.B.; Rowan, E.G.; et al. Action of Varespladib (LY-315920), a Phospholipase A2 Inhibitor, on the Enzymatic, Coagulant and Haemorrhagic Activities of Lachesis muta rhombeata (South-American Bushmaster) Venom. Front. Pharmacol. 2021, 12, 812295.
- Bryan-Quirós, W.; Fernández, J.; Gutiérrez, J.M.; Lewin, M.R.; Lomonte, B. Neutralizing properties of LY315920 toward snake venom group I and II myotoxic phospholipases A2. Toxicon 2019, 157, 1–7.
This entry is offline, you can click here to edit this entry!
