Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Genetics & Heredity
In livestock breeding, the number of vertebrae has gained significant attention due to its impact on carcass quality and quantity. Variations in vertebral traits have been observed across different animal species and breeds, with a strong correlation to growth and meat production. Furthermore, vertebral traits are classified as quantitative characteristics. Molecular marker techniques, such as marker-assisted selection (MAS), have emerged as efficient tools to identify genetic markers associated with vertebral traits.
- livestock
- vertebral traits
- meat production
- genetic markers
1. Introduction
The meat production industry encounters challenges in augmenting carcass and meat quality attributes, significantly influencing consumer preferences and demand amid the global upsurge in meat consumption [1,2]. Key attributes encompassing body morphology, skeletal architecture, vertebral characteristics, muscular development, tenderness, and adipose deposition in livestock species hold paramount significance in governing both meat quality and quantity [3,4,5,6,7,8,9].
In the domain of livestock breeding, the pursuit of augmenting traits related to body size has emerged as a central objective, particularly in China, due to its substantial influence on meat production and carcass traits [10,11,12]. Recent research underscores the economic significance of vertebral count in determining carcass length, weight, and body size traits [10,11,12,13]. Variations in body size have emerged within and between domestic animal species or breeds during livestock evolution [14]. Notably, the number of thoracolumbar vertebrae plays a crucial role in carcass characteristics, particularly carcass length [15]. Across different pig breeds [16], sheep breeds [17], donkeys [10,18], and yaks [19], variations in the number of thoracolumbar vertebrae have been observed. This trait has been considered a selection criterion in commercial animal breeding due to its strong correlations with growth and meat production, exhibiting high heritability (0.60–0.62) and a positive correlation with body length [15].
China has made significant advancements in livestock genetic improvement, particularly in pig breeding [20]. However, enhancing breeding efficiency and accuracy remains a challenge. Molecular marker techniques, such as marker-assisted selection (MAS), have emerged as a rapid and effective approach [21,22,23]. MAS relies on the linkage between phenotypic traits and molecular markers within the genome, enabling the swift and accurate identification of individuals with desired traits, thus improving breeding efficiency. Identifying candidate genes and their polymorphisms associated with vertebral traits holds scientific and economic significance, providing markers for genetic enhancement in livestock.
The influence of genetic factors and signaling pathways on multi-vertebrae traits in livestock is a complex and multifaceted area of research. Several studies have shed light on the genetic underpinnings of these traits, with a focus on genes such as PLAG1, VRTN, PRKG2, MMP4, NR6A1, LTBP2, DCAF7, NCAPG-LCORL, ActRIIB, and TGFβ3, as well as their associations with specific vertebral characteristics in various livestock species, especially pigs, donkeys, and sheep. Consistently, Yan et al. [10] and Liu Z et al. [18] emphasized the quantitative nature of these traits, indicating that they are influenced by multiple genes and intricate biological signaling pathways. This highlights the polygenic nature of multi-vertebrae traits, where numerous genetic factors interplay to determine the final outcome. The genes PLAG1 and NCAPG-LCORL, originally known for their roles in human height, carcass weight, and body length, have emerged as key players in livestock vertebral traits, underscoring their pleiotropic effects [24]. Furthermore, specific mutations in genes like ActRIIB and TGFβ3 have been linked to variations in vertebral number in sheep and pigs, respectively. Liu J et al. [25] identified a point mutation in intron 4 of the ActRIIB gene in Small-Tailed Han sheep associated with vertebral number variation. Similarly, Yue J et al. [26] reported that the mutation g.105179474 G > A in the TGFβ3 gene was associated with rib and thoracolumbar vertebrae numbers in pigs, highlighting the genetic diversity that can underlie these traits. In Beijing Black pigs, Niu N et al. [27] identified a significant link between variant-g.19034 A > C of the VRTN gene and multiple thoracic vertebrae numbers, further exemplifying the genetic complexity of vertebral traits in sheep and pigs.
Donkeys are a comparatively less explored area in this context and have shown promising insights. Shi et al. [28] reported significant associations between specific genetic loci, HOXC8 g.15179224C > T and g.15179674G > A, with lumbar vertebrae length and the number of lumbar vertebrae. This highlights the potential for genetic selection and breeding strategies to influence vertebral traits in donkeys, similar to other livestock species. Nevertheless, the existing body of literature to date has exclusively focused on the investigation of genetic markers associated with vertebral traits in donkeys, sheep, and pigs. However, it is noteworthy that comprehensive investigations into the association of genetic markers with vertebral traits in cattle and horses have been absent from the existing body of research. Drawing upon the aforementioned evidence, it becomes evident that variations in vertebral traits among livestock species (donkeys, sheep, and pigs) can significantly influence changes in body size, carcass length, and weight. Furthermore, it is essential to recognize that vertebral traits are inherently quantitative in nature, controlled by a complex interplay of multiple genes. Thus, the exploration and screening of genes associated with these vertebral traits hold substantial potential for enhancing meat production within the livestock sector, thereby making a notable contribution to the meat industry.
2. Genetic Markers Associated with Number of Vertebrae in Pigs, Donkeys, and Sheep
2.1. Genetic Markers Associated with Number of Vertebrae in Pigs
Pig vertebral classification comprises five distinct segments: cervical, thoracic, lumbar, sacral, and caudal vertebrae. The precise count of cervical, sacral, and caudal vertebrae in pigs is consistent at seven, four, and five, respectively [15]. The key constituents of the vertebral column are the thoracic and lumbar vertebrae, exhibiting considerable variability in their numbers. In Western modern breeds, the thoracic vertebral number spans from 13 to 17, while the lumbar vertebral number ranges from five to seven [13,15]. Wild boars possess 19 thoracic-lumbar vertebrae, whereas Chinese indigenous breeds exhibit a total thoracic and lumbar vertebral count ranging from 19 to 20 [13,15]. Notably, Western commercial breeds such as Large White, Duroc, and Landrace feature a higher number of thoracic-lumbar vertebrae (n = 21 to 23) due to rigorous selective breeding [13,15]. In pigs, some causal or tightly linked genes affecting crucial vertebral traits and used in practical production have been reported in previous studies [29,30,31,32]. In practical pig production, the integration of genetic knowledge regarding vertebral traits allows producers to make informed breeding decisions. Selecting breeding stock based on favorable genetic markers can lead to more robust, efficient, and economically viable pig populations. In addition, vertebral malformations can lead to structural deformities, reduced growth rates, and increased susceptibility to injuries. This, in turn, contributes to the sustainability and competitiveness of the swine industry. The summary of genes associated with vertebral traits is provided in Table 1.
Table 1. Summary of genes associated with vertebral traits in pigs.
| Genes | Associated Traits | Breeds | Country | Reference |
|---|---|---|---|---|
| RSAD2-CMPK2, COL3A1 |
|
Meishan pigs | China | [30] |
| HMGA1, VRTN, BMP2 |
|
Duroc × Landrace × Yorkshire crossbred pigs | [31,32] | |
| TIMP2, EML1, SMN1 |
|
Pigs | [33] | |
| NR6A1, LTBP2 |
|
Xiang pigs | [34] | |
| GREB1L, ABCD4, VRTN, MIB1 |
|
Beijing Black pigs | [27] | |
| ABCD4 Hox family genes (HOXB 1–7, 9, and 13), NTRK2 |
|
Beijing Black pigs | [35] | |
| NR6A1, VRTN PLAG1, BMP2 MC4R |
|
Shanxia Black pigs | [36] | |
| HOXA10 |
|
Pigs | [37] | |
| BMP2 |
|
Duroc × (Landrace × Yorkshire) hybrid pigs | [38] | |
| VRTN, LTBP2, BMPR1A, FOS |
|
White × Minzhu crossbred pigs | [39] | |
| VRTN |
|
Sujiang, Meishan, Bama, Erhualian, and Tibetan pigs | [40] | |
| VRTN |
|
Suhuai pigs | [13] | |
| VRTN |
|
Pigs | [41] | |
| VRTN |
|
Duroc, Landrace, and Large White pigs | Norway | [42] |
| MMP9, VEGF |
|
Pigs | USA | [43] |
| NR6A1 |
|
Large White × Minzhu pigs | China | [44] |
| VRTN, FOS, PROX2, TGFB3 |
|
2.2. Genetic Markers Associated with Number of Vertebrae in Donkeys
The study by Liu Z et al. [18], which involved a comprehensive survey of 455 donkeys, highlighted the presence of diverse configurations of thoracic and lumbar vertebrae in the Dezhou donkey population [18]. These configurations were identified as T18L5, T18L6, T17L6, T17L5, and T19L5. Notably, T18L5 was the most prevalent, accounting for 75.8% of the population. This finding suggests a certain level of variability in vertebral numbers among Dezhou donkeys. Moreover, research conducted by Liu Z et al. also established a correlation between the body size and weight of donkeys and the number of vertebrae (Figure 1) [18,45]. This correlation could potentially open up avenues for selective breeding and manipulation of vertebral numbers to enhance the structural characteristics of donkeys within the industry. However, it is important to note that the genetic mechanisms governing these traits require further investigation.
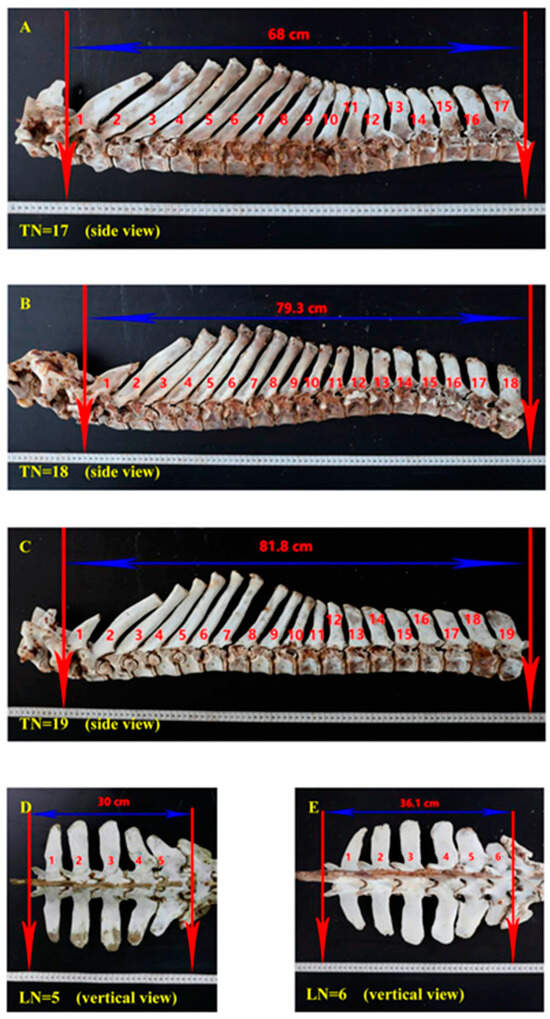
Figure 1. Association of number of vertebrae with body size traits. (A) Side overview of 17 thoracic vertebrae; (B) Eighteen thoracic vertebrae side overview of seventeen; (C) Side overview of 19 thoracic vertebrae; (D) Five lumber vertebrae vertical overview; (E) Six lumber vertebrae vertical overview.
In the context of understanding the genetic basis of vertebral number determination in donkeys, recent studies have made significant contributions [12,18,45,46]. Specifically, the PRKG2 gene has emerged as a key player associated with both the number and length of thoracic and lumbar vertebrae in donkeys [12]. Specific genetic variants within PRKG2 were found to be significantly correlated with thoracic and lumbar vertebrae numbers as well as their length. This genetic insight provides valuable information for further research into the molecular mechanisms behind vertebral development.
It is worth noting that PRKG2, located on chromosome 3, consists of 18 exons and 17 introns in donkeys, as outlined by Wang C et al. [22]. Interestingly, PRKG2 has also been implicated in dwarfism in various species, including American Angus cattle [47], humans [48,49], and dogs [50], underscoring its importance in skeletal development across different organisms. Additionally, its involvement in adipocyte and osteoblast differentiation in the human body, as reported by Yi et al., highlights the multifaceted nature of PRKG2’s functions [51]. In conclusion, the research on vertebral variations in Dezhou donkeys, coupled with the identification of the PRKG2 gene and its associated genetic variants, provides a foundation for further exploration into the genetic mechanisms governing vertebral development. The summary of genes associated with vertebral traits in donkeys is provided in Table 2.
Table 2. Summary of genes associated with vertebral traits in donkeys.
| Genetic Markers | Biological Effect | Breed | Country | Reference |
|---|---|---|---|---|
| DCAF7 |
|
Dezhou donkeys | China | [11] |
| PRKG2 |
|
[12] | ||
| NR6A1 |
|
[18] | ||
| LTBP2 |
|
[46] | ||
| HOXC8 |
|
[28] | ||
| NLGN1, DCC, FBXO4 SLC26A7, TOX, LRP5 WNT7A, LOC123286078, LOC123280142, GABBR2, LOC123277146, LOC123277359, BMP7, B3GAT1, EML2 |
|
[10] |
2.3. Genetic Markers Associated with Number of Vertebrae in Sheep
In the context of sheep anatomy, sheep typically possess a vertebral arrangement comprising seven cervical vertebrae (C), 13 thoracic vertebrae (T), six lumbar vertebrae (L), and four sacral vertebrae (S), totaling 30 vertebrae. Notably, mutations in the thoracolumbar region, such as T14L6 or T13L7, have been identified as the most prevalent [52]. These mutations are associated with multi-vertebrae sheep, which exhibit enhanced adaptability and meat production performance [52]. The cultivation of such multi-spine sheep carries substantial benefits for the economy, society, and ecology, making it a pivotal endeavor for improving the quality and efficiency of the animal husbandry industry.
In the case of Kazakh sheep, an indigenous breed in Western Xinjiang, China, there exists variability in the number of lumbar vertebrae. While most sheep conform to the standard configuration of 13 thoracic vertebrae and six lumbar vertebrae, denoted as T13L6, Kazakh sheep exhibit variations, specifically in T13L7 and T14L6. These variations correspondingly lead to an increase in carcass length by 2.22 cm and 2.93 cm compared to the typical T13L6 Kazakh sheep. Moreover, there is a corresponding elevation in carcass weight by 1.68 kg and 1.90 kg, respectively [53,54,55]. Significant progress has been achieved in China regarding the screening of genetic markers associated with vertebral traits in sheep [56,57,58,59,60,61,62]. For the sake of clarity, we compiled a concise summary of genes related to vertebral count and bone development, presented in Table 3.
Table 3. Summary of genes associated with vertebral traits and bone development in sheep.
| Genes | Associated Traits | Breeds | Country | Reference |
|---|---|---|---|---|
| NR6A1 |
|
Xinjiang Kazakh sheep | China | [56] |
| SFRP4 |
|
Duolang sheep | [57] | |
| SYNDIG1L, UNC13C |
|
Han sheep and Sunite sheep | [52] | |
| TBXT |
|
Sheep | [58] | |
| MGAT4A, KCNH1 CPOX, CPQ |
|
Hu sheep | [59] | |
| LTBP2, SYNDIG1L |
|
Large fat-tailed sheep, Altay sheep, Tibetan sheep |
[60] | |
| VRTN, HoxA |
|
Xinjiang Kazakh sheep | [61] | |
| NDRG2 |
|
Kazakh sheep | [62] | |
| VRTN |
|
China Kazakh sheep | [55] | |
| NID2, ACAN |
|
Afghani sheep | Iran | [63] |
| ALX4, HOXB13, BMP4 EYA2, SULF2 |
|
Ethiopian indigenous sheep | Ethiopia | [64] |
This entry is adapted from the peer-reviewed paper 10.3390/ani14040594
This entry is offline, you can click here to edit this entry!
