Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Psychiatry
Post-traumatic stress disorder (PTSD) is a psychiatric disorder that causes debilitating functional impairment in patients. Observations from survivors of traumatic historical events solidify that this disease is not only associated with personal experiences but can also be inherited from familial traumas. Over the past decades, researchers have focused on epigenetic inheritance to understand how responses to adverse experiences can be passed down to future generations.
- DNA methylation
- histone modification
- intergenerational epigenetic
- maternal inheritance
- non-coding RNA
- paternal inheritance
- transgenerational epigenetic
1. Epigenetic Mechanisms
To promptly react to environmental changes and increase fitness, the expression of genes is under epigenetic control. Epigenetic mechanisms modify the level of transcripts that could subsequently proceed to translation. These changes are flexible yet durable, as they could be changed within a cell cycle and maintained throughout a lifetime [6]. Some of the epigenetic mechanisms commonly used in the human body are DNA methylation, histone modifications, and non-coding RNAs.
1.1. DNA Methylation
DNA methylation is the most studied epigenetic mechanism of all. It was originally discovered in 1944 by Hotchkiss. In the 1980s, it was confirmed that methylated cytosine is involved in gene expression [12]. DNA methylation typically functions as a repressor of transcription of the modified gene, especially when it occurs in the promoter region [13]. It plays a critical role in processes such as silencing retroviral elements, regulating tissue-specific gene expression, imprinting, and X chromosome inactivation [12,14]. The entire process, from placing and removing markers to the recognition of DNA silencing, requires a writer, an eraser, and a reader. The methylation of DNA is regulated and maintained by the DNMT (DNA methyltransferases) family (DNMT, DNMT3a, DNMT3b, and DNMTl) typically on the C5 (carbon number 5) position of cytosines that are followed by a guanine nucleotide; such sites are termed CpG sites [12]. Although the majority of methylations occur on CpG sites, methylations have also been found on cortices non-CpG (CpC, CpA, CpT) sites [12]. The existence of a single demethylation enzyme remains debatable; however, a consensus has been reached on the involvement of Tet enzymes and base excision repair. The ten–eleven translocation (Tet) enzymes (Tet1, Tet2, and Tet3) catalyze the formation of the 5hmC (5-Methylcytosine) intermediate, which could then be converted via further hydroxylation by Tet and deamination by AID/APOBEC to molecules (Thy, 5hmU, 5fC, and 5caC) that will be excised by BER and replaced by a cytosine. Finally, the methylated cytosines are read by the MBD (methyl-CpG binding) proteins, the UHRF (ubiquitin-like, containing PHD and RING finger domains) proteins, and the zinc-finger proteins to silence the expression or reinforce the methylation during replication.
1.2. Histone Modification
Histones are protein molecules that provide the scaffold for DNA to be organized into nucleosomes and later into chromatins, and the modifications on histones will therefore affect chromatin structure as well as the recruitment of nucleosome-remodeling enzymes. Histone modification primarily includes histone (de)acetylation, histone methylation, and histone phosphorylation. Histone acetylation on lysine residues is catalyzed by histone acetyltransferase (HAT). Lysines on both the histone core and tail could be acetylated. Acetylated lysine has a weaker affinity to the DNA molecule, allowing for it to be easily accessible by transcriptosomes for transcription. Deacetylation of lysine by histone deacetylases (HDAC) creates effects opposite to those of histone acetylation, as it condenses the chromatin, repressing gene expression. Lastly, serine, threonine, and tyrosine residues, predominantly those on the histone tail, can undergo histone phosphorylation. A few kinases responsible for histone phosphorylation have been identified, as well as their antagonizing phosphatases [15].
1.3. Non-Coding RNAs (ncRNA)
The final epigenetic processes non-coding RNAs (ncRNA) will be discussed. Only a fraction of the human transcriptome encodes protein [16]. Each class of RNA has unique cellular functions, some of which are types of machinery modulating gene expression levels by chromatin remodeling at the transcriptional or post-transcriptional level [16]. RNAs with epigenetic functions include micro RNA (miRNA), small interfering RNA (siRNA), promoter-associated RNA (PAR), enhancer RNA, and long non-coding RNA [17]. The most abundant ncRNA is the lncRNA. MiRNA and siRNA regulate the expression of around 50% of human genes by binding to and cleaving complementary mRNA transcripts [17]. PAR has been found to have repressive or activation effects in different studies [17]. When associated with the polycomb protein group, PARs exhibit a repressive function. As their name suggests, the enhancer RNAs activate the transcription of genes. LncRNA are the most abundant of all ncRNA, and they recruit chromatin-remodeling proteins [17]. However, since ncRNA asymmetrically segregates into the daughter cells, its role as an epigenetic marker has been under debate [18].
2. Identifying Epigenetic Markers of PTSD
The epigenome-wide association study (EWAS) is the most recent approach to identifying epigenetic markers. Analogous to the genome-wide association study (GWAS) method, which assesses the entire genome for disease association, EWAS aims to assess whole-genome epimutations about diseases, without a hypothesis. Despite many epigenetic mechanisms, due to technological restrictions, most EWAS to date have been focusing on DNA methylations [19]. Compared to GWAS, an ideal EWAS design requires a more deliberate selection of experimental and control samples as well as statistical corrections to account for the dynamic epigenome [20] that varies between populations. The whole-genome data of methylation sites are then obtained from the tissue of interest through various typing and profiling technologies [21], and differences between groups can be compared in a site-specific, CpG-cluster, or regional fashion after the aforementioned statistical corrections [22].
Much like genome-wide association studies, the EWAS method [23] commonly acquires methylome data from peripheral blood samples instead of the tissue of interest (i.e., brain tissue for PTSD), considering the accessibility and simplicity of study design. However, controversy surrounds such a choice of sample tissue, due to the tissue-specific nature of epigenetic markers [20]. All cells in the human body share the same genome, and thus obtaining genomic data from blood samples for GWAS is deemed to be appropriate [20].
Noticeably, the methylome and epigenome do not exhibit uniformity across different tissues [20]. Cells in different tissues perform their distinct tasks and maintain expression patterns through cycles of cell division using epigenetics [20]. Therefore, the resulting data should be interpreted carefully, keeping in mind the epigenetic differences between blood cells and the experiment’s cell of interest [20]. Similarly, the tissue-specific nature of epigenetic markers also requires careful comparison, commonly with statistical analysis, between the acquired data and original epigenetic markings of the discussed tissue to ensure the identified deviation as epimutation [20].
Despite the hardship of conducting epigenetic studies, many advancements have been made in identifying PTSD-related DNA methylation sites. Epigenetic changes in genes participating in physiological processes such as the HPA axis, immune function, neuroplasticity, circadian rhythms, and cell adhesion are of particular interest.
Specifically, PTSD has been linked to dysregulation of the hypothalamus–pituitary–adrenal axis (HPA axis). This stress response pathway develops in early fetal stages [9] and is easily affected by the environment. When the HPA axis is incapable of returning to a normal physiological state, a prolonged state of stress occurs. Among the many genes involved in the HPA axis, two of them are most extensively studied: NCR3C1, the glucocorticoid receptor gene, and FKBP5, the co-chaperone that inhibits glucocorticoid receptor (NR3C1 product) function [24,25,26]. Multiple CpG sites on variants of these genes have been found to contribute to PTSD symptoms and resilience to trauma [26].
Interestingly, methylation on the cg19645279 site on the NR3C1 gene and the cg07485685 site on the FKBP5 is found to be associated with both symptom severity and resilience.
3. Intergenerational and Transgenerational Inheritance of Epigenetic Markers
The identification of epigenetic markers is a big step towards understanding the non-genetic cause of PTSD. Furthermore, genetic or epigenetic changes made in the germline indirectly affect the epigenetics of future generations. Nonetheless, when the epigenetic effect is observed in later generations, it could be passed down via multiple routes which are classified into two categories: intergenerational inheritance and transgenerational inheritance [37]. Even though some researchers consider these two terms to be interchangeable, it is beneficial to differentiate these inheritance processes according to Skinner’s definition. Please note that in the following sections, when referring to transgenerational/intergenerational inheritance studies, corrections will be made according to Skinner’s definition [37].
Intergenerational inheritance implies a transmission process between two generations [38]. As the gamete of the trauma-exposed F0 generation is already present during the exposure, the definition of intergenerational inheritance does not rule out the possibility of direct traumatic exposure of F1 genetic material within the gamete, embryo, and fetus. This means of inheritance is widely studied due to the high malleability of the epigenome during embryogenesis. Transgenerational epigenetic inheritance has a more rigorous criterion based on biological sex and pregnancy status. The affected individual: when the affected individual is male or a non-gestating female, the environmental stressor affects both the individual and their germline cells and, in such cases, the epigenetic change must persist to the third generation to be considered transgenerational inheritance [39]. In gestating females, however, germline cells developing withing within the fetus could also be directly affected by the stimuli [37]. Therefore, any stressor applied is considered a direct stressor to all three generations [40], which renders the next generation to be the first without direct traumatic experience. In short, transgenerational inheritance is only said to occur when the epigenetic marker is present in the first exposure-free generation to the original stressor; this criterion ensures that epigenetic markers are transmitted. For the above reasons, preconception trauma in women, fetoplacental interactions, and improper parental care are considered to be intergenerational inheritance [41] and shall not be confused with the transgenerational inheritance of trauma, as in these scenarios the subject of interest receives direct traumatic experience. The concept of transgenerational, preconceptional, and fetoplacental inheritance is illustrated in Figure 1.
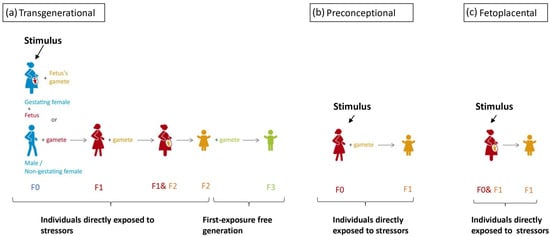
Figure 1. A color-coded schematic drawing of stimulus application timing of each route intergenerational inheritance, with the same color indicating the same individual. (a) For true transgenerational inheritance, stress response should be recorded in the first generation that is not directly affected by the stressor. If a fetus is exposed to stressors during pregnancy, we cannot exclude the epigenetic changes in F1 are independent of changes in F0. Therefore, for male/non-gestating female F0, a response should be recorded in the F2 generation. However, for gestation female F0, stressors directly affected the F0, F1, and F2 generation; therefore, a response would have to be recorded in the F3 generation. (b,c) Preconceptional and fetoplacental inheritance both concerns 2 generations to which the is stressor directly applied, the F0 and the F1 (the gamete or the fetus of F0).
4. Different Timings of Trauma Exposure and Intergenerational Inheritance Studies
There has been compelling evidence suggesting physiological changes in the offspring of parents who experienced traumatic events. In 2018, Yehuda et al. reviewed human studies of the epigenetic intergenerational inheritance of stress [9]. The routes of intergenerational inheritances were categorized by the time of trauma exposure: maternal care (post-natal exposure), fetoplacental interaction (in utero exposure), preconception trauma (gamete exposure), and transgenerational inheritance. Differential findings might suggest that inheritance routes play a significant role in stress inheritance.
4.1. Post-Natal Exposure
Although perturbation caused by insufficient parental care should not be deemed as a means of inheritance, mood disorders can impair the interaction between the patient and their offspring, acting as a form of childhood adversity [42]. Adverse childhood experiences (ACE) have a large impact on the neural development of an individual. ACEs have been associated with psychiatric diseases such as PTSD, anxiety, depression, and bipolar disorder, as well as physical illnesses such as diabetes and cardiovascular diseases. Diseases are caused by differential methylation patterns on genes relating to neurotransmission, specifically the decrease in methylation on FKBP5 (heat shock protein for GR) and MAOA (degradation of monoamine neurotransmitter; also, the increase in methylation on NR3C1 (GR gene), HTR (serotonin receptor), SLC6A4 (serotonin reuptake transporter), and BDNF (promote neuroplasticity)) [43]. On top of these findings, it has been found that maternal overprotection can also cause alterations in hormone expression in holocaust survivors [44]. The complexity of human parenting style has also added to the complication of mood regulation studies.
4.2. In Utero Exposure
Experiences as early as in utero exposure have been found to contribute to PTSD vulnerability through DNA methylation [45]. Increased methylation has been found in the promoter region of the NR3C1/2 gene in both mothers (F0) who were pregnant during the Tutsi genocide and their children (F1) [9,46]. Similar findings have also been found in women pregnant while experiencing domestic violence, in the war zones of the democratic republic of Congo [34], during the Rwanda genocide [46], and the Quebec ice storm. A study on women who experienced sexual violence showed expression in F1 through fetal placental differential methylation patterns on the SLC6A4INJ2, OXTR promoter, and NINJ2 genes and related regions [33].
4.3. Preconceptional Inheritance and Transgenerational Inheritance
Among all routes of epigenetic inheritance of stress, preconceptional trauma inheritance and transgenerational inheritance are particularly under debate [47]. This is based on the understanding that global reprogramming of the epigenome occurs in the mammalian germline immediately after fertilization to erase acquired epigenetic marks or epigenotype [14,48]. Although retention has been found in methylation in some regions of the genome, the sperm histone PTM, and sperm small ncRNAs, more studies are required to understand the mechanism behind the preservation of epigenetic markers [39]. Still, observations of epigenetic changes in later generations provide evidence that some methylations exhibit meiotic stability [49].
This entry is adapted from the peer-reviewed paper 10.3390/life14010098
This entry is offline, you can click here to edit this entry!
