Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Several distinct pathogenic coronaviruses have emerged, including the pandemic SARS-CoV-2, which is difficult to curtail despite the availability of licensed vaccines. The difficulty in managing SARS-CoV-2 is linked to changes in the variants’ proteins, especially in the spike protein (SP) used for viral entry. These mutations, especially in the SP, enable the virus to evade immune responses induced by natural infection or vaccination. However, some parts of the SP in the S1 subunit and the S2 subunit are considered conserved among coronaviruses.
- SARS-CoV-2
- vaccine
- spike protein
- mutation
- conserved epitopes
1. Introduction
In the history of humans, the Coronavirus disease (COVID-19) caused by the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) has made an indelible mark characterized by a contagious respiratory pandemic [1][2]. Consequently, 660 million cases and approximately 6.6 million deaths have been reported as of December 2022 [3]. Since its emergence, studies have been focusing on developing preventive and therapeutic measures against this disease, and several vaccines have been licensed [4][5]. However, the constant mutation of the SARS-CoV-2 virus, which results in different variants, narrows the effectiveness of the licensed vaccines against SARS-CoV-2 infections, thus suggesting a needed continuous effort in developing a SARS-CoV-2 universal vaccine [6]. Besides the pandemic caused by SARS-CoV-2, other human coronaviruses (huCoVs) including human CoV (HCoV)-229E (1962), HCoV-OC43 (1967), SARS-CoV (2002), HCoV-NL63 (2004), HCoV-HKUI (2005), Middle East respiratory syndrome coronavirus (MERS)-CoV (2012) and SARS-CoV-2 (2019) have been implicated in different outbreaks since the start of the 21st century [7][8][9][10][11]. Although the trend of the emergence of coronaviruses remains unclear, adequate preparation must be made for a possible emerging or re-emerging strain. In this case, developing an effective universal vaccine against CoVs should take advantage of the conserved portions of the virus, especially the spike protein (SP).
Levels of similarities have been observed among the emerged CoVs [12][13][14]. For instance, the SARS-CoV-2 viral genome sequence analysis revealed its phylogenetic similarity with SARS-CoV (79%) and MERS-CoV (50%) [15][16][17]. Like other betacoronaviruses, the SARS-CoV-2 genome consists of 27 proteins encoded by 14 open reading frames (ORF), including non-structural proteins (nsp) that are encoded by the ORF 1 and 2 present at the 5′-terminal regions [18][19]. More importantly, the SARS-CoV-2 structural proteins, notably the SP, envelope protein (E), membrane protein (M), nucleocapsid (N) and eight accessory proteins encoded by the 3′-terminal region of the genomes, are in like manner with other betacoronaviruses [19].
Sequence analysis of the SARS-CoV-2 SP revealed a close association with SARS-CoV regarding amino acid composition as well as comparable binding affinity to human angiotensin-converting enzyme 2 (hACE2) [20][21]. The SP, which is a highly N-glycosylated class I transmembrane fusion protein, plays a critical role during coronavirus infection. The SP mediates the attachment of the virus into the cell receptor and facilitates the viral-host membrane fusion [22][23]. The SP also assembles into trimers on the virus surface and cleaves into two subunits, S1 and S2, during the cell infection [24][25]. The large protein S1 domain contains the RBD that is responsible for binding to the host cell receptor and varies extensively in all isolates of the CoVs [20][24][25]. Meanwhile, the S2 domain, which facilitates the virus-cell fusion, is made up of the fusion peptide (FP) and heptad repeat regions (HR1 and HR2), which are conserved among the isolates of the CoVs [26][27], the conserved membrane-proximal external region (MPER) and the transmembrane domain (TM) [28][29][30][31][32][33][34]. After the binding of the S1 RBD domain to the ACE2, the S2 subunit then inserts its FP into the cell membrane leading to the assemblage of the HR1 and HR2 into a six-helix structure to drive the cellular and viral membrane closely together for viral entry (Figure 1) [35]. Notably, the major determinant of cell tropism in most coronaviruses is typically linked to the structure of the SP [21][25][36][37][38][39][40]. Phylogenetic, bioinformatic and homology structural modelling analyses showed that the RBD of SARS-CoV-2 only has 64% shared identity with the SARS-CoV, while the NTD has 51% similarity [41]. However, the study revealed that within the S2, the fusion protein (FP) is 93% identical and the HR1 is 88% identical, while the HR2 and the TM are respectively 100% and 93% similar.
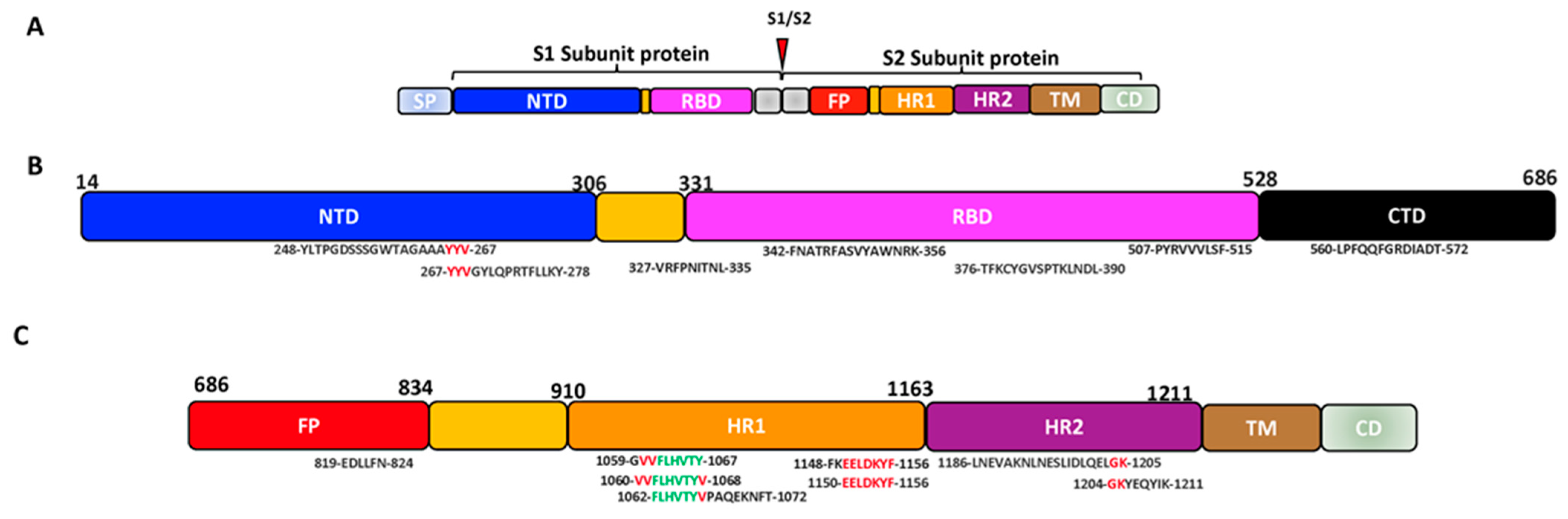
Figure 1. SARS-CoV-2 cell entry into the target cell. (A) SARS-CoV-2 virus with its SP. The SP contains the S1 and the S2 subunit. The S1 subunit comprises the NTD and RBD, while the S2 comprises the FP, HR1 and HR2. (B) The schematic summary of SARS-CoV-2 cell entry. SARS-CoV-2 binds to the host ACE receptor using the RBD domain of the S1 (upper panel). After the activation of the S2, SARS-CoV-2 uses the FP and HR for fusion and gains entry into the host (lower panel) [35][42]. Image created by Biorender.com. Adapted from “SARS-CoV-2 Targeting of ACE2 Receptor and Entry in Infected Cell” and “An In-depth Look into the Structure of the SARS-CoV2 Spike Glycoprotein”, by BioRender.com (Accessed on 22 February 2023). Retrieved from https://app.biorender.com/biorender-templates.
SARS-CoV-2 and other SARS-related coronaviruses (e.g., SARS-CoV) can utilize distinct domains within the S1 subunit to identify different attachment and entry receptors in the host cell surface [20][40]. For instance, the differences in the SPs of the known huCoVs contribute to their difference in pathogenesis and the site of infection (lower or upper respiratory) [36]. Taking a closer look at SARS-CoV-2 as an example, Laporte et al. described that the higher transmissibility experienced with the SARS-CoV-2 compared to other human coronaviruses is due to its abundant replication in the upper respiratory tracts [43]. They mentioned that the SARS-CoV-2 SP has an intrinsic temperature preference of 33 °C, like the temperature of human respiratory tracts, instead of the 37 °C required by other human CoVs. In addition to this, it was revealed that the SARS-CoV-2 has multiple cell entry activators, including TMPRSS2 and TMPRSS13 protease, broadening its tropism. As mentioned by Korber et al., a D614G mutation observed in the SARS-CoV-2 variant S1 also resulted in the wider spread of the virus at different geographical regions with higher viral loads in the respiratory tracts [44]. These differences observed in the SP of the CoVs contribute to the difficulties in developing a sustainable vaccine against the emerging variants of SARS-CoV-2. It is therefore a necessity to develop a vaccine that can prevent the spread of all present or emerging variants of SARS-CoV-2.
2. SARS-CoV-2 S Conserved Regions as a Potential Target for Vaccine Development
Developing a vaccine for viruses with multiple strains or variants tends to take advantage of the conserved epitopes present in the virus. Conserved epitopes are epitopes that are relatively the same among different strains of a pathogen. For example, due to the multiple strains of influenza over the years, the means of developing a universal vaccine has been the use of the conserved epitopes on the hemagglutinin (HA stalk) or the matrix ectodomain (M2e) [45][46][47][48][49][50][51]. The strategy of using conserved epitopes has also been used in the development of preventative vaccines for HIV, dengue virus, Lassa fever virus (LASV), hepatitis virus and Kaposi’s sarcoma-associated herpesvirus (KSHV) [52][53][54][55][56][57][58][59][60]. Therefore, identifying the conserved regions in the SP of SARS-CoV variants will be of great importance in developing a universal vaccine against CoVs. Although mutations in SARS-CoV-2 had a great impact on the SP, studies have revealed that certain conserved epitopes in S1 can induce neutralizing antibodies [61]. Interestingly, some monoclonal antibodies (including 7B11, 18F3, S309 and its Fab, S315, 154C, S304, 240C and VHH-72) that recognize SARS-CoV and MERS-CoV could cross-react and cross-neutralize SARS-CoV-2 by recognizing the ACE2 binding sites on the SARS-CoV-2 RBD [62].
Jaiswal et al. mathematically (in-house developed PERL scripts) revealed sets of epitopes on the S1 subunit around 453 to 538 that interact with the ACE and are 99% conserved among the variants of SARS-CoV-2 [63]. Their study further identified conserved common neutralizing epitopes on the SARS-CoV-2, including YLTPGDSSSGWTAGAAAYYV (247–267 aa), TFKCYGVSPTKLNDL (376–390 aa) on S1 and LNEVAKNLNESLIDLQELGK (1186–1205 aa) on the S2 [63]. A recent immunoinformatic study predicted conserved and highly immunogenic CTL-induced epitopes on S1 VRFPNITNL (327–335 aa) and PYRVVVLSF (507–515 aa) (Table 1), while the CTL-induced epitopes on S2 were identified to be VVFLHVTYV (1060–1068 aa) and GVVFLHVTY (1059–1067 aa) (Table 2) [64]. Based on the conservancy, antigenicity, allergenicity, population coverage and transmembrane location, another study chose potential conserved epitopes from the S1 (FNATRFASVYAWNRK, 342–356 aa) (Table 1), S2 (FLHVTYVPAQEKNFT, 1062–1072 aa) (Table 2) and the E/M protein to construct a SARS-CoV-2 vaccine and revealed that it had high immunogenicity and broad neutralizing activity against the SARS-CoV-2 RBD (Figure 2) [65]. Jiang et al., with web-based analytic tools, also predicted potential T cell epitopes induced by the SARS-CoV-2 SP and narrowed them down to CD4 or CD8 T cell epitopes using ELIspot and a cytolytic assay [66]. In their observation, YYVGYLQPRTFLLKY (264–278 aa), located at the end of the NTD and upstream of the RBD, is highly conserved among 11 variants of SARS-CoV-2 VOCs and variants of interest (VOIs) and could induce the T cells. Further studies also suggested that these epitopes could be well recognized by most HLA alleles globally [66].
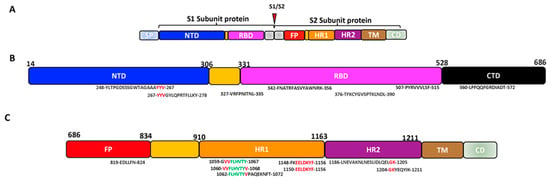
Figure 2. The schematic representation of some possible conserved epitopes on the SARS-CoV-2 SP (Black: positions occurring in a set of predicted epitopes; Red: positions occurring in 2 sets of predicted epitopes; Green: positions present in 3 sets of predicted epitopes). (A) Schematic structure of the SARS-CoV-2 spike protein. (B) Structure of the SARS-CoV-2 S1 subunit showing the conserved epitopes on the NTD and RBD. (C) Structure of the SARS-CoV-2 S2 subunit showing the conserved epitopes on the FP and HR.
Table 1. Some predicted conserved epitopes on the SARS-CoV-2 S1 subunit that could induce neutralizing antibodies and/or adaptive immune responses.
| Conserved Epitopes | Position | Immune Response Induced | Type of Study | Refs. | |
|---|---|---|---|---|---|
| B cells/Neutralizing antibodies | T cells | ||||
| YLTPGDSSSGWTAGAAAYYV | 248–267 aa | Yes | Yes | Mathematically (in-house developed PERL scripts), in vivo | [63][67] |
| YYVGYLQPRTFLLKY | 264–278 aa | NT | Yes | Web-based analytic tools | [66] |
| VRFPNITNL | 327–335 aa | NT | Yes | Immunoinformatic, in vivo | [64][68] |
| FNATRFASVYAWNRK | 342–356 aa | Yes | Yes | In silico, T-cell epitope mapping, molecular dynamics simulations, immunoinformatic | [65][69][70] |
| TFKCYGVSPTKLNDL | 376–390 aa | Yes | Yes | Mathematically (in-house developed PERL scripts), bioinformatics, monoclonal antibody targeting | [63][71] |
| PYRVVVLSF | 507–515 aa | NT | Yes | Immunoinformatic | [64] |
| LPFQQFGRDIADT | 560–572 aa | Yes | Yes | PepSeq Analysis | [72] |
NT—Not tested.
Table 2. Some predicted conserved epitopes on the SARS-CoV-2 S2 subunit that could induce neutralizing antibodies and/or adaptive immune responses.
| Conserved Epitopes | Position | Immune Response Induced | Type of Study | Refs. | |
|---|---|---|---|---|---|
| Neutralizing antibodies | T cells | ||||
| EDLLFN | 819–824 aa | Yes | NT | Epitope-resolved profiling, structural and functional test | [72][73] |
| EELDKYF | 1150–1156 aa | Yes | NT | Epitope-resolved profiling, structural and functional | [72][73] |
| GVVFLHVTY | 1059–1067 aa | NT | Yes | Immunoinformatic | [64][74] |
| VVFLHVTYV | 1060–1068 aa | NT | Yes | Immunoinformatic | [64][74] |
| FLHVTYVPAQEKNFT | 1062–1072 aa | Yes | Yes | In silico, in vivo | [65] |
| SPDVDLGDISGINAS | 1161–1175 aa | Yes | NT | In vivo | [75] |
| LNEVAKNLNESLIDLQELGK | 1186–1205 aa | Yes | Yes | Mathematically (in-house developed PERL scripts), bioinformatic, in vivo | [63][67][76] |
| GKYEQYIK | 1204–1211 aa | NT | NT | Antiviral inhibitory activity | [34] |
NT—Not tested.
Considering the role played by the S2 subunit during SARS-CoV-2 infection, it could also be targeted to induce immune responses against SARS-CoV-2. Interestingly, Ladner et al. generated an epitope-resolved analysis of IgG cross-reactivity among all CoVs in COVID-19-negative and recovered patients using a highly multiplexed peptide assay (PepSeq) and discovered that the epitopes at the FP, which is 93% similar among strains of betacoronaviruses and alphacoronaviruses, produced broadly neutralizing antibodies against the endemic coronaviruses, including SARS-CoV, MERS-CoV and SARS-CoV-2 [41][72]. Likewise, the HR2 region of the S2 is 100% conserved among the variants of the SARS-CoV-2 [41][75]. The S2 subunit proteins could also induce cross-reactive antibodies against the SARS-CoV SP and the endemic CoVs [77]. The S2 has been reported to induce neutralizing antibodies or T cell responses targeting the FP proximal region and HR2 domain of S2 in COVID-19 patients or animals vaccinated or infected with different CoVs [78][79][80][81]. Interestingly, due to prior population exposure to common cold coronaviruses, nAbs and memory B and T cells against SARS-CoV-2 were found in some individuals who have never been infected by SARS-CoV-2 [82][83]. Pre-existing antibodies against conserved epitopes of S2, such as residues 901–906, 810–816, 851–856, 1040–1044 and 1205–1212, showed the greatest cross-reactivity and hindered SARS-CoV-2 entry into cells [82]. Other identified regions that are most widely recognized among SARS-CoV-2 linear epitopes in convalescent donors are EELDKYF (1150–1156 aa) within the stem helix of the HR2 terminal and EDLLFN (819–824 aa), which overlaps the FP and is adjacent to the S2 cleavage [72][73]. Moreover, a study conducted by Song et al. revealed that monoclonal antibody CC9.3 isolated from individuals before SARS-CoV-2 infection was characterized to recognize the S2 subunit of the SARS-CoV-2 and other huCoVs [84].
From the sera of patients recovered from SARS-CoV-2, Pinto et al. isolated five monoclonal antibodies that could recognize the stem-helix (SH) of the S2 subunits of other betacoronaviruses including the OC43 strain [73]. In addition, Lu et al. identified and crystallized T cell follicle helper cells (cTfh) among patients who recovered from the mild symptoms of COVID-19 and revealed that these cTfh could recognize SARS-CoV-2 S2 subunit epitopes (864–882 aa) that are conserved among the emerging variants [85]. In a recent publication, Wu et al. identified a monoclonal antibody hMab5.17 that could recognize the SARS-CoV-2 HR2 domain that is adjacent to TM (SPDVDLGDISGINAS; 1161–1175 aa) and could protect against SARS-CoV-2 in the Syrian hamster. They further cloned the mAb hMab5.17 and demonstrated that it could neutralize SARS-CoV-2 variants [75].
Another highly conserved region located at the S2 region is the SARS-CoV-2 MPER-like region (MPER) (GKYEQYIK; 1204–1211 aa), which has great potential for being used as an antigen for broadly neutralizing antibodies (bnAbs) [34][86][87]. Yu et al. have demonstrated that a cholesterol-conjugated lipopeptide containing SARS-CoV-2 MPER prepared by using cholesteryl succinate monoester could inhibit viral entry, indicating the importance of MPER in viral entry and fusion [34]. Like the MPER of HIV-1, the MPER of SARS-CoV-2 could possibly induce bnAbs against the variants of SARS-CoV-2. Studies have shown that bnAbs 4E10, 2F5, 10E8 and LN01 could interact with the MPER of the HIV-1 gp41 to prevent infection [88][89][90]. Likewise, the SARS-CoV-2 MPER could be a suitable immunogen for inducing neutralizing antibodies [34][87].
The above findings suggested that the S2 subunit of the SARS-CoV-2 is conserved among the previous strains of human coronaviruses and the SARS-CoV-2 variants and can induce cross-reactive antibodies.
This entry is adapted from the peer-reviewed paper 10.3390/vaccines11030545
References
- Gralinski, L.E.; Menachery, V.D. Return of the Coronavirus: 2019-nCoV. Viruses 2020, 12, 135.
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506.
- Worldometer. Coronavirus Disease. Available online: https://www.google.com/search?q=coronavirus+death+toll&rlz=1C5CHFA_enCA1017CA1017&oq=coronavirus+death&aqs=chrome.0.0i131i433i512j69i57j0i512l8.6462j1j4&sourceid=chrome&ie=UTF-8-colocmid=/m/02j71&coasync=0 (accessed on 29 September 2022).
- Lopez Bernal, J.; Andrews, N.; Gower, C.; Gallagher, E.; Simmons, R.; Thelwall, S.; Stowe, J.; Tessier, E.; Groves, N.; Dabrera, G. Effectiveness of COVID-19 vaccines against the B. 1.617. 2 (Delta) variant. N. Engl. J. Med. 2021, 385, 585–594.
- Kumar, S.; Saurabh, M.K.; Maharshi, V. Efficacy and safety of potential vaccine candidates against coronavirus disease 2019: A systematic review. J. Adv. Pharm. Technol. Res. 2021, 12, 215.
- Zhao, F.; Zai, X.; Zhang, Z.; Xu, J.; Chen, W. Challenges and developments in universal vaccine design against SARS-CoV-2 variants. Npj Vaccines 2022, 7, 1–12.
- Van Den Brand, J.M.; Smits, S.L.; Haagmans, B.L. Pathogenesis of Middle East respiratory syndrome coronavirus. J. Pathol. 2015, 235, 175–184.
- Mackay, I.M.; Arden, K.E. MERS coronavirus: Diagnostics, epidemiology and transmission. Virol. J. 2015, 12, 1–21.
- Alharbi, N.K.; Kulkarni, S.S.; Falzarano, D. Immune Responses to MERS-CoV in Humans and Animals. In Microbial Pathogenesis; Springer: Berlin/Heidelberg, Germany, 2021; pp. 85–97.
- Cui, J.; Li, F.; Shi, Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192.
- Cueno, M.E.; Imai, K. Structural comparison of the SARS-CoV 2 spike protein relative to other human-infecting coronaviruses. Front. Med. 2021, 7, 594439.
- Salian, V.S.; Wright, J.A.; Vedell, P.T.; Nair, S.; Li, C.; Kandimalla, M.; Tang, X.; Carmona Porquera, E.M.; Kalari, K.R.; Kandimalla, K.K. COVID-19 transmission, current treatment, and future therapeutic strategies. Mol. Pharm. 2021, 18, 754–771.
- Petrosillo, N.; Viceconte, G.; Ergonul, O.; Ippolito, G.; Petersen, E. COVID-19, SARS and MERS: Are they closely related? Clin. Microbiol. Infect. 2020, 26, 729–734.
- Chen, Y.; Liu, Q.; Guo, D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020, 92, 418–423.
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.S.; de Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.L.; Lauber, C.; Leontovich, A.M.; Neuman, B.W. Severe acute respiratory syndrome-related coronavirus: The species and its viruses–a statement of the Coronavirus Study Group. Nat. Microbiol. 2020, 5, 536–544.
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574.
- Beyer, D.K.; Forero, A. Mechanisms of antiviral immune evasion of SARS-CoV-2. J. Mol. Biol. 2022, 434, 167265.
- Wu, A.; Peng, Y.; Huang, B.; Ding, X.; Wang, X.; Niu, P.; Meng, J.; Zhu, Z.; Zhang, Z.; Wang, J. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 2020, 27, 325–328.
- Malik, Y.S.; Sircar, S.; Bhat, S.; Sharun, K.; Dhama, K.; Dadar, M.; Tiwari, R.; Chaicumpa, W. Emerging novel coronavirus (2019-nCoV)—Current scenario, evolutionary perspective based on genome analysis and recent developments. Vet. Q. 2020, 40, 68–76.
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220.
- Li, Q.; Wu, J.; Nie, J.; Zhang, L.; Hao, H.; Liu, S.; Zhao, C.; Zhang, Q.; Liu, H.; Nie, L. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell 2020, 182, 1284–1294.e1289.
- Ao, Z.; Ouyang, M.J.; Olukitibi, T.A.; Yao, X. SARS-CoV-2 Delta spike protein enhances the viral fusogenicity and inflammatory cytokine production. Iscience 2022, 25, 104759.
- Daniloski, Z.; Jordan, T.X.; Ilmain, J.K.; Guo, X.; Bhabha, G.; Sanjana, N.E. The Spike D614G mutation increases SARS-CoV-2 infection of multiple human cell types. Elife 2020, 10, e65365–e65379.
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020, 181, 281–292.e286.
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454.
- Reguera, J.; Mudgal, G.; Santiago, C.; Casasnovas, J.M. A structural view of coronavirus–receptor interactions. Virus Res. 2014, 194, 3–15.
- Tang, T.; Bidon, M.; Jaimes, J.A.; Whittaker, G.R.; Daniel, S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antivir. Res. 2020, 178, 104792.
- Poston, D.; Weisblum, Y.; Wise, H.; Templeton, K.; Jenks, S.; Hatziioannou, T.; Bieniasz, P. Absence of severe acute respiratory syndrome coronavirus 2 neutralizing activity in prepandemic sera from individuals with recent seasonal coronavirus infection. Clin. Infect. Dis. 2021, 73, e1208–e1211.
- Elko, E.A.; Nelson, G.A.; Mead, H.L.; Kelley, E.J.; Carvalho, S.T.; Sarbo, N.G.; Harms, C.E.; Le Verche, V.; Cardoso, A.A.; Ely, J.L. COVID-19 vaccination elicits an evolving, cross-reactive antibody response to epitopes conserved with endemic coronavirus spike proteins. Cell Rep. 2022, 40, 111022.
- Grobben, M.; van der Straten, K.; Brouwer, P.J.; Brinkkemper, M.; Maisonnasse, P.; Dereuddre-Bosquet, N.; Appelman, B.; Lavell, A.A.; van Vught, L.A.; Burger, J.A. Cross-reactive antibodies after SARS-CoV-2 infection and vaccination. Elife 2021, 10, e70330.
- Millet, J.K.; Whittaker, G.R. Physiological and molecular triggers for SARS-CoV membrane fusion and entry into host cells. Virology 2018, 517, 3–8.
- Chambers, P.; Pringle, C.R.; Easton, A.J. Heptad repeat sequences are located adjacent to hydrophobic regions in several types of virus fusion glycoproteins. J. Gen. Virol. 1990, 71, 3075–3080.
- Xia, S.; Xu, W.; Wang, Q.; Wang, C.; Hua, C.; Li, W.; Lu, L.; Jiang, S. Peptide-based membrane fusion inhibitors targeting HCoV-229E spike protein HR1 and HR2 domains. Int. J. Mol. Sci. 2018, 19, 487.
- Yu, D.; Zhu, Y.; Jiao, T.; Wu, T.; Xiao, X.; Qin, B.; Chong, H.; Lei, X.; Ren, L.; Cui, S. Structure-based design and characterization of novel fusion-inhibitory lipopeptides against SARS-CoV-2 and emerging variants. Emerg. Microbes Infect. 2021, 10, 1227–1240.
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.-Y. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 2020, 181, 894–904.e899.
- Masters, P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006, 66, 193–292.
- Glowacka, I.; Bertram, S.; Müller, M.A.; Allen, P.; Soilleux, E.; Pfefferle, S.; Steffen, I.; Tsegaye, T.S.; He, Y.; Gnirss, K. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011, 85, 4122–4134.
- Lu, G.; Hu, Y.; Wang, Q.; Qi, J.; Gao, F.; Li, Y.; Zhang, Y.; Zhang, W.; Yuan, Y.; Bao, J. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature 2013, 500, 227–231.
- Wang, N.; Shi, X.; Jiang, L.; Zhang, S.; Wang, D.; Tong, P.; Guo, D.; Fu, L.; Cui, Y.; Liu, X. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013, 23, 986–993.
- Hulswit, R.J.; Lang, Y.; Bakkers, M.J.; Li, W.; Li, Z.; Schouten, A.; Ophorst, B.; Van Kuppeveld, F.J.; Boons, G.-J.; Bosch, B.-J. Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A. Proc. Natl. Acad. Sci. USA 2019, 116, 2681–2690.
- Jaimes, J.A.; André, N.M.; Chappie, J.S.; Millet, J.K.; Whittaker, G.R. Phylogenetic analysis and structural modeling of SARS-CoV-2 spike protein reveals an evolutionary distinct and proteolytically sensitive activation loop. J. Mol. Biol. 2020, 432, 3309–3325.
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280.e278.
- Laporte, M.; Raeymaekers, V.; Van Berwaer, R.; Vandeput, J.; Marchand-Casas, I.; Thibaut, H.-J.; Van Looveren, D.; Martens, K.; Hoffmann, M.; Maes, P. The SARS-CoV-2 and other human coronavirus spike proteins are fine-tuned towards temperature and proteases of the human airways. PLoS Pathog. 2021, 17, e1009500.
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B. Tracking changes in SARS-CoV-2 spike: Evidence that D614G increases infectivity of the COVID-19 virus. Cell 2020, 182, 812–827.e819.
- Erbelding, E.J.; Post, D.J.; Stemmy, E.J.; Roberts, P.C.; Augustine, A.D.; Ferguson, S.; Paules, C.I.; Graham, B.S.; Fauci, A.S. A universal influenza vaccine: The strategic plan for the National Institute of Allergy and Infectious Diseases. J. Infect. Dis. 2018, 218, 347–354.
- Nachbagauer, R.; Krammer, F. Universal influenza virus vaccines and therapeutic antibodies. Clin. Microbiol. Infect. 2017, 23, 222–228.
- Neirynck, S.; Deroo, T.; Saelens, X.; Vanlandschoot, P.; Jou, W.M.; Fiers, W. A universal influenza A vaccine based on the extracellular domain of the M2 protein. Nat. Med. 1999, 5, 1157–1163.
- Saelens, X. The role of matrix protein 2 ectodomain in the development of universal influenza vaccines. J. Infect. Dis. 2019, 219, S68–S74.
- Uranowska, K.; Tyborowska, J.; Jurek, A.; Szewczyk, B.a.; Gromadzka, B. Hemagglutinin stalk domain from H5N1 strain as a potentially universal antigen. Acta Biochim. Pol. 2014, 61, 541–550.
- Turley, C.B.; Rupp, R.E.; Johnson, C.; Taylor, D.N.; Wolfson, J.; Tussey, L.; Kavita, U.; Stanberry, L.; Shaw, A. Safety and immunogenicity of a recombinant M2e–flagellin influenza vaccine (STF2. 4xM2e) in healthy adults. Vaccine 2011, 29, 5145–5152.
- Olukitibi, T.; Ao, Z.; Azizi, H.; Mahmoudi, M.; Coombs, K.; Kobasa, D.; Kobinger, G.; Yao, X. Development and characterization of influenza M2 ectodomain and/or hemagglutinin stalk-based dendritic cell-targeting vaccines. Front. Microbiol. 2022, 13, 937192–937206.
- Chauhan, V.; Rungta, T.; Goyal, K.; Singh, M.P. Designing a multi-epitope based vaccine to combat Kaposi Sarcoma utilizing immunoinformatics approach. Sci. Rep. 2019, 9, 1–15.
- Guest, J.D.; Pierce, B.G. Structure-based and rational design of a hepatitis C virus vaccine. Viruses 2021, 13, 837.
- Ahmed, S.F.; Quadeer, A.A.; Barton, J.P.; McKay, M.R. Cross-serotypically conserved epitope recommendations for a universal T cell-based dengue vaccine. PLoS Negl. Trop. Dis. 2020, 14, e0008676.
- Dixit, N.K. Design of Monovalent and Chimeric Tetravalent Dengue Vaccine Using an Immunoinformatics Approach. Int. J. Pept. Res. Ther. 2021, 27, 2607–2624.
- Ali, M.; Pandey, R.K.; Khatoon, N.; Narula, A.; Mishra, A.; Prajapati, V.K. Exploring dengue genome to construct a multi-epitope based subunit vaccine by utilizing immunoinformatics approach to battle against dengue infection. Sci. Rep. 2017, 7, 1–13.
- Sampath, A.; Padmanabhan, R. Molecular targets for flavivirus drug discovery. Antivir. Res. 2009, 81, 6–15.
- Mateo, M.; Reynard, S.; Journeaux, A.; Germain, C.; Hortion, J.; Carnec, X.; Picard, C.; Baillet, N.; Borges-Cardoso, V.; Merabet, O. A single-shot Lassa vaccine induces long-term immunity and protects cynomolgus monkeys against heterologous strains. Sci. Transl. Med. 2021, 13, eabf6348.
- ter Meulen, J.; Badusche, M.; Satoguina, J.; Strecker, T.; Lenz, O.; Loeliger, C.; Sakho, M.; Koulemou, K.; Koivogui, L.; Hoerauf, A. Old and New World arenaviruses share a highly conserved epitope in the fusion domain of the glycoprotein 2, which is recognized by Lassa virus-specific human CD4+ T-cell clones. Virology 2004, 321, 134–143.
- ter Meulen, J.; Badusche, M.; Kuhnt, K.; Doetze, A.; Satoguina, J.; Marti, T.; Loeliger, C.; Koulemou, K.; Koivogui, L.; Schmitz, H. Characterization of human CD4+ T-cell clones recognizing conserved and variable epitopes of the Lassa virus nucleoprotein. J. Virol. 2000, 74, 2186–2192.
- Sankaranarayanan, S.; Mohkhedkar, M.; Janakiraman, V. Mutations in spike protein T cell epitopes of SARS-COV-2 variants: Plausible influence on vaccine efficacy. Biochim. Et Biophys. Acta (BBA)-Mol. Basis Dis. 2022, 1868, 166432.
- Kombe Kombe, A.J.; Zahid, A.; Mohammed, A.; Shi, R.; Jin, T. Potent molecular feature-based neutralizing monoclonal antibodies as promising therapeutics against SARS-CoV-2 infection. Front. Mol. Biosci. 2021, 8, 670815.
- Jaiswal, V.; Lee, H.-J. Conservation and Evolution of Antigenic Determinants of SARS-CoV-2: An Insight for Immune Escape and Vaccine Design. Front. Immunol. 2022, 13, 832106.
- Bagherzadeh, M.A.; Izadi, M.; Baesi, K.; Jahromi, M.A.M.; Pirestani, M. Considering epitopes conservity in targeting SARS-CoV-2 mutations in variants: A novel immunoinformatics approach to vaccine design. Sci. Rep. 2022, 12, 1–17.
- Kibria, K.; Faruque, M.; Islam, M.; Ullah, H.; Mahmud, S.; Miah, M.; Saleh, A.A. A conserved subunit vaccine designed against SARS-CoV-2 variants showed evidence in neutralizing the virus. Appl. Microbiol. Biotechnol. 2022, 106, 4091–4114.
- Jiang, S.; Wu, S.; Zhao, G.; He, Y.; Guo, X.; Zhang, Z.; Hou, J.; Ding, Y.; Cheng, A.; Wang, B. Identification of a promiscuous conserved CTL epitope within the SARS-CoV-2 spike protein. Emerg. Microbes Infect. 2022, 11, 730–740.
- Vishwakarma, P.; Yadav, N.; Rizvi, Z.A.; Khan, N.A.; Chiranjivi, A.K.; Mani, S.; Bansal, M.; Dwivedi, P.; Shrivastava, T.; Kumar, R. Severe acute respiratory syndrome coronavirus 2 spike protein based novel epitopes induce potent immune responses in vivo and inhibit viral replication in vitro. Front. Immunol. 2021, 12, 613045.
- Muraoka, D.; Situo, D.; Sawada, S.-i.; Akiyoshi, K.; Harada, N.; Ikeda, H. Identification of a dominant CD8+ CTL epitope in the SARS-associated coronavirus 2 spike protein. Vaccine 2020, 38, 7697–7701.
- Lin, J.; Law, R.; Korosec, C.S.; Zhou, C.; Koh, W.H.; Ghaemi, M.S.; Samaan, P.; Ooi, H.K.; Yue, F.; Gingras, A.-C. Longitudinal Assessment of SARS-CoV-2 Specific T Cell Cytokine-Producing Responses for 1 Year Reveals Persistence of Multi-Cytokine Proliferative Responses, with Greater Immunity Associated with Disease Severity. Int. J. Mol. Sci. 2022, 23, 4341.
- Martin, W.R.; Cheng, F. A rational design of a multi-epitope vaccine against SARS-CoV-2 which accounts for the glycan shield of the spike glycoprotein. J. Biomol. Struct. Dyn. 2021, 40, 7099–7113.
- Lim, H.X.; Masomian, M.; Khalid, K.; Kumar, A.U.; MacAry, P.A.; Poh, C.L. Identification of B-Cell Epitopes for Eliciting Neutralizing Antibodies against the SARS-CoV-2 Spike Protein through Bioinformatics and Monoclonal Antibody Targeting. Int. J. Mol. Sci. 2022, 23, 4341.
- Ladner, J.T.; Henson, S.N.; Boyle, A.S.; Engelbrektson, A.L.; Fink, Z.W.; Rahee, F.; D’ambrozio, J.; Schaecher, K.E.; Stone, M.; Dong, W. Epitope-resolved profiling of the SARS-CoV-2 antibody response identifies cross-reactivity with endemic human coronaviruses. Cell Rep. Med. 2021, 2, 100189.
- Pinto, D.; Sauer, M.M.; Czudnochowski, N.; Low, J.S.; Tortorici, M.A.; Housley, M.P.; Noack, J.; Walls, A.C.; Bowen, J.E.; Guarino, B. Broad betacoronavirus neutralization by a stem helix–specific human antibody. Science 2021, 373, 1109–1116.
- Mallavarpu Ambrose, J.; Priya Veeraraghavan, V.; Kullappan, M.; Chellapandiyan, P.; Krishna Mohan, S.; Manivel, V.A. Comparison of immunological profiles of SARS-CoV-2 variants in the COVID-19 pandemic trends: An immunoinformatics approach. Antibiotics 2021, 10, 535.
- Wu, W.-L.; Chiang, C.-Y.; Lai, S.-C.; Yu, C.-Y.; Huang, Y.-L.; Liao, H.-C.; Liao, C.-L.; Chen, H.-W.; Liu, S.-J. Monoclonal antibody targeting the conserved region of the SARS-CoV-2 spike protein to overcome viral variants. JCI Insight 2022, 7, e157597.
- Cuspoca, A.F.; Estrada, P.I.; Velez-van-Meerbeke, A. Molecular mimicry of SARS-CoV-2 spike protein in the nervous system: A bioinformatics approach. Comput. Struct. Biotechnol. J. 2022, 20, 6041–6054.
- Ng, K.T.; Mohd-Ismail, N.K.; Tan, Y.-J. Spike S2 subunit: The dark horse in the race for prophylactic and therapeutic interventions against SARS-CoV-2. Vaccines 2021, 9, 178.
- Li, Y.; Lai, D.-y.; Zhang, H.-n.; Jiang, H.-w.; Tian, X.; Ma, M.-l.; Qi, H.; Meng, Q.-f.; Guo, S.-j.; Wu, Y.; et al. Linear epitopes of SARS-CoV-2 spike protein elicit neutralizing antibodies in COVID-19 patients. Cell. Mol. Immunol. 2020, 17, 1095–1097.
- Poh, C.M.; Carissimo, G.; Wang, B.; Amrun, S.N.; Lee, C.Y.-P.; Chee, R.S.-L.; Fong, S.-W.; Yeo, N.K.-W.; Lee, W.-H.; Torres-Ruesta, A.; et al. Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralising antibodies in COVID-19 patients. Nat. Commun. 2020, 11, 2806.
- Xia, S.; Yan, L.; Xu, W.; Agrawal, A.S.; Algaissi, A.; Tseng, C.K.; Wang, Q.; Du, L.; Tan, W.; Wilson, I.A.; et al. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci. Adv. 2019, 5, eaav4580.
- Smith, T.R.F.; Patel, A.; Ramos, S.; Elwood, D.; Zhu, X.; Yan, J.; Gary, E.N.; Walker, S.N.; Schultheis, K.; Purwar, M.; et al. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat. Commun. 2020, 11, 2601.
- Ng, K.W.; Faulkner, N.; Cornish, G.H.; Rosa, A.; Harvey, R.; Hussain, S.; Ulferts, R.; Earl, C.; Wrobel, A.G.; Benton, D.J. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science 2020, 370, 1339–1343.
- Nguyen-Contant, P.; Embong, A.K.; Kanagaiah, P.; Chaves, F.A.; Yang, H.; Branche, A.R.; Topham, D.J.; Sangster, M.Y. S protein-reactive IgG and memory B cell production after human SARS-CoV-2 infection includes broad reactivity to the S2 subunit. MBio 2020, 11, e01991-20.
- Song, G.; He, W.-t.; Callaghan, S.; Anzanello, F.; Huang, D.; Ricketts, J.; Torres, J.L.; Beutler, N.; Peng, L.; Vargas, S. Cross-reactive serum and memory B-cell responses to spike protein in SARS-CoV-2 and endemic coronavirus infection. Nat. Commun. 2021, 12, 2938.
- Lu, X.; Hosono, Y.; Nagae, M.; Ishizuka, S.; Ishikawa, E.; Motooka, D.; Ozaki, Y.; Sax, N.; Maeda, Y.; Kato, Y. Identification of conserved SARS-CoV-2 spike epitopes that expand public cTfh clonotypes in mild COVID-19 patients. J. Exp. Med. 2021, 218, e20211327.
- Shah, M.; Woo, H.G. Omicron: A heavily mutated SARS-CoV-2 variant exhibits stronger binding to ACE2 and potently escapes approved COVID-19 therapeutic antibodies. Front. Immunol. 2022, 12, 6031.
- Zhu, Y.; Yu, D.; Hu, Y.; Wu, T.; Chong, H.; He, Y. SARS-CoV-2-derived fusion inhibitor lipopeptides exhibit highly potent and broad-spectrum activity against divergent human coronaviruses. Signal Transduct. Target. Ther. 2021, 6, 294.
- Pinto, D.; Fenwick, C.; Caillat, C.; Silacci, C.; Guseva, S.; Dehez, F.; Chipot, C.; Barbieri, S.; Minola, A.; Jarrossay, D. Structural basis for broad HIV-1 neutralization by the MPER-specific human broadly neutralizing antibody LN01. Cell Host Microbe 2019, 26, 623–637.e628.
- Zhang, L.; Irimia, A.; He, L.; Landais, E.; Rantalainen, K.; Leaman, D.P.; Vollbrecht, T.; Stano, A.; Sands, D.I.; Kim, A.S. An MPER antibody neutralizes HIV-1 using germline features shared among donors. Nat. Commun. 2019, 10, 1–16.
- Pietzsch, J.; Scheid, J.F.; Mouquet, H.; Seaman, M.S.; Broder, C.C.; Nussenzweig, M.C. Anti-gp41 antibodies cloned from HIV-infected patients with broadly neutralizing serologic activity. J. Virol. 2010, 84, 5032–5042.
This entry is offline, you can click here to edit this entry!
