Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Breast cancer (BC) is the most common kind of cancer in women, accounting for around 30% of all new cancer diagnoses; it is also the second most fatal malignancy after lung and bronchial cancers. Centered on deep convolutional neural networks, a new BC histopathological image category blind inpainting convolutional neural network (BiCNN) model has been developed. It was developed to cope with the two-class categorization of BC on the diagnostic image.
- breast cancer diagnosis
- deep mutual learning
- histopathology imaging diagnosis
1. Introduction
Breast cancer (BC) is the most common kind of cancer in women, accounting for around 30% of all new cancer diagnoses; it is also the second most fatal malignancy after lung and bronchial cancers [1]. According to the most recent data from the International Agency for Research on Cancer, which is part of the World Health Organization, breast cancer has exceeded lung cancer as the most frequent cancer, with 2.26 million new cases in 2020, overtaking lung cancer. It presents a major threat to the lives and health of women. Early diagnosis is crucial in the fight against cancer, and this can only be achieved with a reliable detection system. Two techniques that have been developed to aid in the diagnosis of breast cancer are medical image processing and digital pathology, respectively [2][3][4]. BC has two particularly alarming features among the many forms of cancer: being the most frequent disease in women across the globe and having a much higher fatality rate than additional kinds of cancer, because the histopathological examination is the most often utilized approach for the diagnosis of breast cancer. In many cases, pathologists still use the visual evaluation of histological samples beneath the microscope to make a diagnosis. Automated histopathological image classification is a study area that might speed up and reduce the risk of mistakes in BC diagnosis [5]. Histopathology uses a biopsy to obtain images of the diseased tissue [6][7]. Early identification is significant for illness treatment and a safer prognosis [8]. Noninvasive BC screening procedures include clinical breast assessment and tomography tests, such as magnetic resonance, ultrasound, and mammography. However, the requirement for verifying the identification of BC is the pathological study of a slice of the suspicious region by a diagnostician. Glass slides tarnished with hematoxylin and eosin are used to examine the microscopic details of the questionable tissue [9]. There are various analytical modes used for BC detection. Some of the general modes are mammography, magnetic resonance imaging (MRI), positron emission tomography (PET), breast ultrasound, surgery, or fine-needle aspiration to target the nerve of the alleged area (histopathological images), etc., as shown in Figure 1 [10][11].
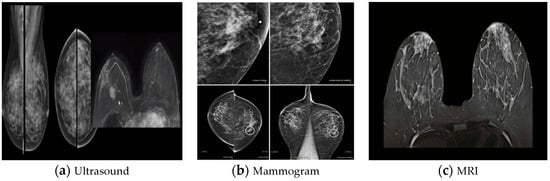
Figure 1. Medical imaging modalities for breast tissue: (a) ultrasound, (b) mammogram, and (c) MRI [10].
Different methods, such as rule-based and machine-learning approaches, are used to evaluate breast cancer digital pathology images [12]. Recently, it has been shown that deep-learning-based approaches, which automate the whole processing, outperform classical machine-learning techniques in numerous image-assessment tasks [13]. Successful applications of convolutional neural networks (CNNs) in medical imaging have allowed for the early diagnosis of diabetic retinopathy, the prediction of bone disease and age, and other problems. Earlier deep-learning-based functions in histological microscopic image processing have demonstrated their capacity to be effective in the detection of breast cancer. Machine learning has played an increasingly important role in breast cancer detection over the last several decades. Several probabilistic, statistical, and optimization strategies could be used in the machine-learning approach to derive a classification model from a dataset [14].
Breast carcinoma is a commonly classified histopathology established on the selection of morphological aspects of the cancers, with 20 main cancer categories and 18 lesser subtypes. Invasive ductal carcinoma (IDC) and invasive lobular carcinoma (ILC) are the two primary histological groups of breast cancer, with approximately 70–80% of all cases falling into one of these categories [15][16]. Deep-learning (DL) methods are capable of autonomously extracting features, retrieving information from data, and learning sophisticated abstract interpretations of the data. DL techniques are powerful. They can resolve typical feature-extraction issues and have found use in a selection of sectors, including computer vision and biomedicine.
Centered on deep convolutional neural networks, a new BC histopathological image category blind inpainting convolutional neural network (BiCNN) model has been developed. It was developed to cope with the two-class categorization of BC on the diagnostic image. The BiCNN model uses previous knowledge of the BC class and subclass labels to constrain the distance between the characteristics of distinct BC pathology images [17]. A data-augmented technique is provided to suit the acceptance of whole-slide image identification [18]. The transfer-fine-tuning training approach is employed as an appropriate training approach [19] to increase the accuracy of BC histological image categorization. Figure 2 and Figure 3 demonstrate some of the finer characteristics of the pathological images of BC. Samples (a) through (e) in Figure 2 are all ductal carcinomas (DCs). The phyllodes tumor is sample (f). The colors and forms of the cells in samples (a)–(e) all belong to DCs, even though they are all DC samples. Samples (e) and (f) have a striking resemblance in terms of color and cell shape; however, they are classified as distinct classes. Figure 3 depicts abnormal images at various magnification levels. There is a substantial variance in the visual features across the various magnifications, even though they are all from the same subject [20].
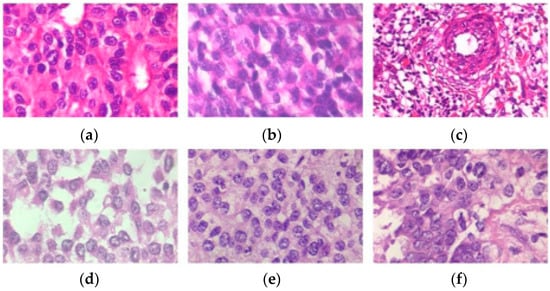
Figure 2. Samples (a–e) are ductal carcinomas (DCs), while sample (f) is a phyllodes tumor carcinoma (PTC) from a woman with breast cancer. Each image is a 400× magnification from the BreakHis archive.
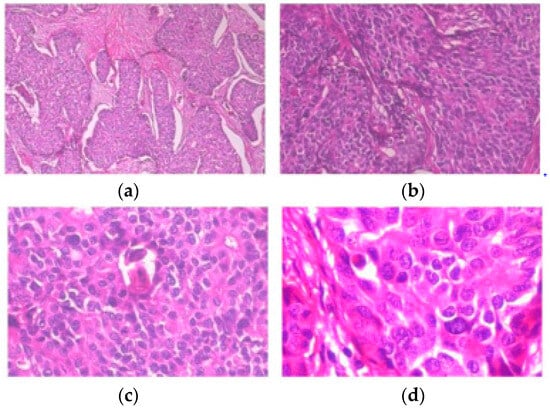
Figure 3. Slides of breast ductal carcinoma from the same patient at (a) 40×, (b) 100×, (c) 200×, and (d) 400×. BreakHis has provided the images [20].
2. Histopathological Image Diagnosis for Breast Cancer Diagnosis Based on Deep Mutual Learning
The National Institute of Oncology in Rabat, Morocco, received 116 surgical breast specimens with invasive cancer of an unknown nature, resulting in 328 digital slides. These photos were properly classified into one of three types: normal tissue–benign lesions, in situ cancer, or aggressive carcinoma. It was shown that, despite the small size of the dataset, the classification model developed in this research was able to accurately predict the likelihood of a BC diagnosis [21]. To compare the performance of chronic myelogenous leukemia (CML)- and DL-based techniques, the author also provided a visual analysis of the histological results to categorize breast cancer. CML-based approaches utilize three feature extractors to extract hand-crafted features and combine them to build an image representation for five traditional classifiers. The DL-based techniques utilized the well-known VGG-19 DL design, which was fine-tuned using histopathological images. The data showed that the DL methods outperformed the CML methods, with an accuracy range of 94.05 to 98.13% for the binary classification and 76.77 to 88.95% for the eighth-class classification [22]. The DCNN-based heterogeneous ensemble method for mitotic nuclei identification was used for breast histopathology images using the DHE-Mit-Classifier. Histopathological biopsy samples were examined for the presence of mitotic patches, and the DHE-Mit-Classifier was used to sort them into mitotic and nonmitotic nuclei. A heterogeneous ensemble was constructed using five independent DCNNs. the mitotic nuclei’s structural, textural, and morphological characteristics remain captured by these DCNNs, which included a variety of architectural styles. The recommended ensemble had an F-score of 0.77, a recall of 0.71, a precision of 0.83, and an area under the curve (AUC) accuracy–recall of 0.83, which surpassed the test set of 0.80. The F-score and accuracy indicated that this ensemble might be utilized to build a pathologist’s helper [23]. The BC patients benefited from the enhanced and multiclass whole-slide imaging (WSI) segmentation uses of the CNN. These components organize information collected from CNNs into pathologists’ predictions. Pathologists need instruments that can speed up the time to perform histological analyses, provide a second opinion, or even point out areas of concern during routine screening. This yielded a sensitivity of 90.77%, a precision of 91.27%, an F1 score of 84.17%, and a specificity of 94.03%. The area subdivision module acquired a sensitivity of 71.83%, an IOU of 88.23%, an intersection over union (IOU) of 93.43%, a precision of 96.10%, an F1 score of 82.94%, a specificity of 96.19%, and an AUC of 0.88 for the improved WSI segmentation [24]. A hybrid model based on DCNNs and pulse-coupled neural networks (PCNNs) was developed. Transfer learning (TL) was used in this study due to the necessity for huge datasets to train and tune the CNNs, which were not accessible for medical images. TL can be an efficient method when dealing with tiny datasets. The document’s application was assessed using three public standard datasets, DDMS, INbreast, and BCDR, for the instruction and analysis, and MIAS for testing alone. The findings demonstrated the benefit of combining the PCNN with the CNN over other approaches for the same public datasets. The hybrid model accurately predicted DDMS (98.72%), BCDR (96.94%), and breast cancer (97.5%). The proposed hybrid model was tested on a previously unreported MIAS dataset and showed an accuracy of 98.7%. In the Results section, further assessment measures can be found [25]. There are a variety of digital pathology image-evaluation techniques for breast cancer, including rule-based and machine-learning approaches [26]. Lately, DL-based processes have been proven to outpace traditional machine-learning techniques in several image-evaluation tasks, computerizing the whole-processing process [27]. Convolution neural networks (CNNs) have been utilized effectively in the medical imaging field to detect diabetic retinopathy, forecast bone disease and age, and other issues. Earlier DL-based functions in histological microscopic image processing have shown their ability to be useful in the diagnosis of breast cancer. The detection of BC has become more dependent on machine learning over the last several decades. The machine-learning method includes a variety of probabilistic, statistical, and optimization techniques for deriving a classification model from a dataset [28].
This entry is adapted from the peer-reviewed paper 10.3390/diagnostics14010095
References
- Siegel, R.L.; Miller, K.D.; Fedewa, S.A.; Ahnen, D.J.; Meester, R.G.; Barzi, A.; Jemal, A. Colorectal cancer statistics, 2017. CA Cancer J. Clin. 2017, 67, 177–193.
- Spanhol, F.A.; Oliveira, L.S.; Cavalin, P.R.; Petitjean, C.; Heutte, L. Deep features for breast cancer histopathological image classification. In Proceedings of the 2017 IEEE International Conference on Systems, Man and Cybernetics (SMC), Banff, AB, Canada, 5–8 October 2017; pp. 1868–1873.
- Desai, M.; Shah, M. An anatomization on breast cancer detection and diagnosis employing multi-layer perceptron neural network (MLP) and Convolutional neural network (CNN). Clin. eHealth 2021, 4, 1–11.
- Alanazi, S.A.; Kamruzzaman, M.M.; Sarker, N.I.; Alruwaili, M.; Alhwaiti, Y.; Alshammari, N.; Siddiqi, M.H. Boosting Breast Cancer Detection Using Convolutional Neural Network. J. Health Eng. 2021, 2021, 5528622.
- Lakhani, S.R.; Ellis, I.O.; Schnitt, S.; Tan, P.; van de Vijver, M. WHO Classification of Tumors of the Breast, 4th ed.; WHO Press: Geneva, Switzerland, 2012; Volume 4.
- Das, A.; Devarampati, V.K.; Nair, M.S. NAS-SGAN: A Semi-Supervised Generative Adversarial Network Model for Atypia Scoring of Breast Cancer Histopathological Images. IEEE J. Biomed. Health Inform. 2021, 26, 2276–2287.
- Liu, Y.; Li, A.; Liu, J.; Meng, G.; Wang, M. TSDLPP: A Novel Two-Stage Deep Learning Framework for Prognosis Prediction Based on Whole Slide Histopathological Images. IEEE/ACM Trans. Comput. Biol. Bioinform. 2021, 19, 2523–2532.
- World Health Organization. Cancer. 2019. Available online: https://www.who.int/health-topics/cancer (accessed on 15 June 2022).
- Bejnordi, B.E.; Zuidhof, G.; Balkenhol, M.; Hermsen, M.; Bult, P.; van Ginneken, B.; Karssemeijer, N.; Litjens, G.; van der Laak, J. Context-aware stacked convolutional neural networks for classification of breast carcinomas in whole-slide histopathology images. J. Med. Imaging 2017, 4, 044504.
- Houssein, E.H.; Emam, M.M.; Ali, A.A.; Suganthan, P.N. Deep and machine learning techniques for medical imaging-based breast cancer: A comprehensive review. Expert Syst. Appl. 2021, 167, 114161.
- Houssein, E.H.; Emam, M.M.; Ali, A.A. An efficient multilevel thresholding segmentation method for thermography breast cancer imaging based on improved chimp optimization algorithm. Expert Syst. Appl. 2021, 185, 115651.
- Zebari, D.A.; Ibrahim, D.A.; Zeebaree, D.Q.; Haron, H.; Salih, M.S.; Damaševičius, R.; Mohammed, M.A. Systematic Review of Computing Approaches for Breast Cancer Detection Based Computer Aided Diagnosis Using Mammogram Images. Appl. Artif. Intell. 2021, 35, 2157–2203.
- Gupta, I.; Gupta, S.; Singh, S. Different CNN-based Architectures for Detection of Invasive Ductal Carcinoma in Breast Using Histopathology Images. Int. J. Image Graph. 2021, 21, 2140003.
- Ogundokun, R.O.; Misra, S.; Douglas, M.; Damaševičius, R.; Maskeliūnas, R. Medical Internet-of-Things Based Breast Cancer Diagnosis Using Hyperparameter-Optimized Neural Networks. Futur. Internet 2022, 14, 153.
- Motlagh, M.H.; Jannesari, M.; Aboulkheyr, H.; Khosravi, P.; Elemento, O.; Totonchi, M.; Hajirasouliha, I. Breast cancer histopathological image classification: A deep learning approach. bioRxiv 2018.
- Sharma, S.; Mehra, R. Conventional Machine Learning and Deep Learning Approach for Multi-Classification of Breast Cancer Histopathology Images—A Comparative Insight. J. Digit. Imaging 2020, 33, 632–654.
- Wei, B.; Han, Z.; He, X.; Yin, Y. Deep learning model-based breast cancer histopathological image classification. In Proceedings of the 2017 IEEE 2nd International Conference on Cloud Computing and Big Data Analysis (ICCCBDA), Chengdu, China, 28–30 April 2017; pp. 348–353.
- Oyewola, D.O.; Dada, E.G.; Misra, S.; Damaševičius, R. A Novel Data Augmentation Convolutional Neural Network for Detecting Malaria Parasite in Blood Smear Images. Appl. Artif. Intell. 2022, 36, 2033473.
- Diwakaran, M.; Surendran, D. Breast Cancer Prognosis Based on Transfer Learning Techniques in Deep Neural Networks. Inf. Technol. Control. 2023, 52, 381–396.
- Li, L.; Pan, X.; Yang, H.; Liu, Z.; He, Y.; Li, Z.; Fan, Y.; Cao, Z.; Zhang, L. Multi-task deep learning for fine-grained classification and grading in breast cancer histopathological images. Multimed. Tools Appl. 2020, 79, 14509–14528.
- El Agouri, H.; Azizi, M.; El Attar, H.; El Khannoussi, M.; Ibrahimi, A.; Kabbaj, R.; Kadiri, H.; BekarSabein, S.; EchCharif, S.; El Khannoussi, B. Assessment of deep learning algorithms to predict histopathological diagnosis of breast cancer: First Moroccan prospective study on a private dataset. BMC Res. Notes 2022, 15, 66.
- Said, B.; Liu, X.; Wan, Y.; Zheng, Z.; Ferkous, C.; Ma, X.; Li, Z.; Bardou, D. Conventional machine learning versus deep learning for magnification dependent histopathological breast cancer image classification: A comparative study with a visual explanation. Diagnostics 2021, 11, 528.
- Sohail, A.; Khan, A.; Nisar, H.; Tabassum, S.; Zameer, A. Mitotic nuclei analysis in breast cancer histopathology images using deep ensemble classifier. Med. Image Anal. 2021, 72, 102121.
- Zeiser, F.A.; da Costa, C.A.; Ramos, G.d.O.; Bohn, H.C.; Santos, I.; Roehe, A.V. DeepBatch: A hybrid deep learning model for interpretable diagnosis of breast cancer in whole-slide images. Expert Syst. Appl. 2021, 185, 115586.
- Altaf, M.M. A hybrid deep learning model for breast cancer diagnosis based on transfer learning and pulse-coupled neural networks. Math. Biosci. Eng. 2021, 18, 5029–5046.
- Robertson, S.; Azizpour, H.; Smith, K.; Hartman, J. Digital image analysis in breast pathology—From image processing techniques to artificial intelligence. Transl. Res. 2017, 1931–5244.
- Ching, T.; Himmelstein, D.S.; Beaulieu-Jones, B.K.; Kalinin, A.A.; Do, B.T.; Way, G.P.; Ferrero, E.; Agapow, P.-M.; Xie, W.; Rosen, G.L.; et al. Opportunities and Obstacles for Deep Learning in Biology and Medicine. bioRxiv 2017.
- Singh, S.; Kumar, R. Histopathological image analysis for breast cancer detection using cubic SVM. In Proceedings of the 2020 7th International Conference on Signal Processing and Integrated Networks (SPIN), Noida, India, 27–28 February 2020; pp. 498–503.
This entry is offline, you can click here to edit this entry!
