Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Powdery mildew (PM) is one of the most common Cannabis sativa diseases. In spite of this, very few documented studies have characterized the resistance genes involved in PM defense mechanisms, or sources of natural genetic resistance in cannabis.
- Cannabis sativa
- powdery mildew
- mildew resistance locus o
- nucleotide-binding and leucine-rich repeat receptors
- disease resistance genes
- broad-spectrum resistance
1. Introduction
Plant diseases caused by pathogenic fungi, oomycetes, bacteria and viruses lead to yield losses, reducing their quality and economic value. These losses can be heavy; for instance, they can reach ~40% in rice and maize [1].
Powdery mildew (PM) is one of the most common plant diseases, caused by several fungi taxa belonging to the Erysiphales order of the Ascomycota phylum, which infects a wide range of plant species [2][3].
In contrast to well-known mycelial fungal/oomycete root rot pathogens, like Fusarium or Pythium, these biotrophic plant pathogens only infect plant tissues growing out of the ground, and the lower leaves are generally the most affected, with only their epidermal cell layer targeted [3]. In a susceptible host plant, the fungal conidium germinates, penetrates the cell wall and establishes a specialized structure, referred to as ‘haustorium’, to absorb nutrients [4]. Then, surface hyphae develop, as well as reproductive structures and new spores, resulting into an extensive surficial hyphal network. As the disease progresses, the PM may spread up and down the length of the crop.
PM fungi grow well with high humidity levels and a moderate temperature, thus greenhouses conditions provide an ideal temperate environment for the spread of the infection, representing a great issue in breeding programs [5]. The disease also has a significant impact on plant growth and yield quality. For instance, a reduction of up to 25% in grain yield has been observed in susceptible wheat cultivars [6].
Asexual reproduction is the predominant strategy to generate PM fungi. The lifestyle of these organisms is a relevant issue for molecular investigations; in fact, efforts to establish a reliable protocol for the stable transformation of PM fungi have often been hampered by the difficulty to cultivate them in vitro [3], and many aspects of their biology have not been completely elucidated. However, several PM fungi genomes have been sequenced, for instance, those associated with barley, wheat, pea and Arabidopsis hosts [3].
Cannabis sativa belongs to the Cannabaceae family and is a dicotyledonous plant which is increasingly cultivated all over the world, due to its adaptability to a wide range of environmental conditions [7]. It is used as a source of industrial fiber, seed oil and food, as well as for health and recreational purposes [8]. The increase in cannabis breeding has led to a massive pathogen exposure, resulting in diseases playing a crucial role in its production. In spite of the availability of its genome sequences, few research works have investigated the pathogen defense mechanisms from a molecular point of view, as well as the underlying genetic and metabolic pathways [9].
Cannabis is susceptible to PM disease [10], which can reduce its yield and photosynthesis rate by damaging foliage and preventing the light from reaching its surface, resulting in premature plant senescence. PM represents a relevant limitation for cannabis production [11][12], and the economic impact of this disease has not yet been precisely evaluated in this crop [13].
The use of pesticides against PM in cannabis could have health risks for the consumer, and alternative methods include environmental control and applications of rhizobacteria promoting plant growth [14][15]. Currently, several products to manage PM in cannabis are available, like the bio-fungicide Regalia Maxx (an extract of giant knotweed) [15] and lacto-fermented products [15], such as Cyclone. Despite these pest management strategies, PM is still one of the most relevant biological diseases for cannabis, and the discovery and characterization of PM resistance genes is crucial for improving the cannabis industry in a sustainable way [10].
Resistance PM genes were found in hops (Humulus lupulus), the most closely related species to C. sativa [16][17]. In cannabis, despite a wide range of diseases being reported, very few documented R genes are known [9]. Emerging molecular studies have reported two primary mechanisms for qualitative resistance against PM in cannabis, but only recently: gene-for-gene resistance [18] and mlo-based resistance [13][19].
The first mechanism occurs when a pathogen-secreted effector protein is recognized by the compatible protein generated by the plant host resistance (R) genes, which are often characterized by conserved nucleotide-binding site (NBS) and/or leucine-rich repeat (LRR) domains (also termed NLRs) [20][21][22]. NLRs, whose mechanisms have been increasingly understood in recent years, are immune receptors and key components of the plant innate immune system, on which plants rely for defense against pathogen infections [23]. They represent the major class of intracellular innate immune receptors and the most represented group of resistance genes. To date, several NBS–LRR resistance genes and quantitative trait loci (QTLs) for plant resistance to pathogens were mapped in plants, some of which were also cloned [24][25], and, in many cases, a co-localization between QTLs and genes was highlighted. This made it possible to identify candidate genes and to develop molecular markers for plant resistance [24][25]. In cannabis, the involvement of NLRs in gene-for-gene interaction with PM has been recently demonstrated [18].
The second mechanism involves loss-of-function mutations of susceptibility (S) genes. The Mildew resistance locus o (MLO) genes are a family of S genes encoding seven transmembrane domain proteins only found in plants, thus helping the infection spread when interacting with PM fungi [26][27]. Their overexpression results in an enhanced susceptibility to PM [28]. Conversely, their loss-of-function mutations (mlo) seem to be a reliable way to protect plants from the infection, and they have a greater potential for durable PM resistance than R-gene resistance, which can be overcome more easily by new pathogen races [29]. Furthermore, mlo-based resistance is commonly non-race-specific and, as a consequence, is effective against the vast majority of PM isolates [30]. mlo-based resistance was initially observed in barley [31], and subsequently many researchers focused their efforts on understanding the molecular mechanisms behind it, discovering the broad-spectrum resistance (BSR) peculiarity in barley, and extending their research to other plant species [32].
2. Broad-Spectrum Disease Resistance and NLR- and mlo-Based Mechanisms
BSR confers resistance against more than one pathogen species (species-nonspecific) or against most races belonging to the same species (race-nonspecific) [33][34]. It is usually durable, remaining effective for long periods, even though the plant is exposed to the pathogen while still growing [33][34].
Most R genes are able to confer high levels of race-specific resistance against a single pathogen, even though some genes, such as those belonging to the wall-associated kinase (WAK) family, were found to be non-race-specific broad spectrum resistance genes [35]. However, due to mutations and virulence variations in pathogens, the effectiveness of the R genes is generally not very durable [34]. Conversely, the partial resistance regulated by QTLs is commonly race-nonspecific, although, in most cases, it provides an insufficient defense against pathogen attacks [34]. Combining R genes and QTLs is an effective strategy for disease control but may be technically challenging and requires a lot of time [34].
Given the above, BSR is a desirable trait and the selection of new cultivars with BSR characteristics has become a crucial crop breeding aim.
Most BSR genes have been reported to encode pattern recognition receptors (PRRs), as well as defense-signaling and pathogenesis-related proteins (PRs) [34]. NLR proteins also mediate defense mechanisms against broad spectrum of pathogens [34][36][37][38], even though they may become ineffective due to virulence variations in pathogens.
Furthermore, several S genes, whose loss-of-function mutations decrease the compatibility between pathogens and plant hosts, have been investigated and identified as BSR genes [34].
2.1. Nucleotide-Binding and Leucine-Rich Repeat Receptors and Their Role in the Immune System
The plant innate immune system consists of two layers: the first one includes the recognition of pathogen-associated molecular patterns (PAMPs) by membrane-associated PRRs, which activate PAMP-triggered immunity (PTI) [39][40]. The second layer results from the recognition of pathogen avirulence (Avr) effectors, leading to an effective and race-specific effector-triggered immunity (ETI), which is generally able to control specific pathogen attacks [20][23]. The ETI response mainly involves the nucleotide-binding and leucine-rich repeat receptors (NLRs) and other cytoplasmic proteins [36][38][41]. Both PRR and NLR-triggered immunity (NTI) lead to a downstream defense response, including the production of reactive oxygen species (ROS), a flux of extracellular calcium, kinase activation and transcriptional regulation in order to combat the infection [37][42]. ROS generation in response to the perception of the pathogen typically culminates in a hypersensitive response (HR) in many resistant genotypes, resulting in localized and very rapid cell death at the infection site [43]. Several transcription factor families, such as AP2/ERF, bHLH, MYB, NAC, WRKY and bZIP [44][45], can be involved in this immune response. After the immune recognition, defense signaling propagates to tissues distant from those where the infection occurred. Defense intensity and duration can be different between PTI and NTI [46]. NLRs induce a stronger and longer defense response over time, which often leads to a programmed cell death [21][37].
NLRs consist of a central NB domain, including the conserved P-loop motif required for ATP/ADP binding and NLR activity [47], and a C-terminal LRR, which is highly polymorphic and confers NLR recognition specificity [48]. NLRs are classified into two subgroups, according to their N-terminal domain: TIR-NB-LRR (TNL) and CC-NB-LRR (CNL) proteins, characterized by a Toll-like and a coiled-coil domain, respectively [24].
NLRs can be located in different subcellular organelles and districts, such as the cytoplasm, nucleus, plasma membrane and endoplasmic reticulum [37][49]. In plant genomes, they can be found either as isolated genes or organized in clusters, enabling the evolution of immune receptors [20][49]. More specifically, many NLRs, named sensor NLRs, perceive pathogen effectors, while others, referred to as helper NLRs, assist immune signaling [21]. NLRs can also be organized in networks, in which several helper NLRs act as signaling hubs for sensor NLRs and other immune receptors, which are localized on the plant cell surface. Pathogens primarily attempt to suppress NLR networks, facilitating the spread of the infection; thus, a deep understanding of the network interaction mechanisms could help to prevent plant disease [21].
NLRs were found to confer disease resistance against PM in many plant species. For instance, the mildew locus a (Mla) NLR gene has been demonstrated to be responsible for resistance against diverse fungal pathogens in cereal crops. In barley, Mla locus confers specific isolate immunity against the PM fungus Blumeria graminis f. sp. hordei (Bgh), and it has been proved that LRRs are largely responsible for the recognition specificity of structurally related effectors by MLAs [50], suggesting that MLA receptors may be driven in the Bgh recognition effectors by the presence of a common structural effector scaffold [50].
Regarding BSR genes encoding NLRs, the first identified species-nonspecific BSR NLR proteins were found in Arabidopsis resistance against two bacteria, Ralstonia solanacearum and Pseudomonas syringae, working synergically as a dual R-gene system [51]. Recently it was demonstrated in Nicotiana benthamiana that NLR proteins recognize the effectors of Pseudomonas and Xanthomonas species [52].
NLR-based resistance mechanisms have been the subject of several investigations to date [21].
2.2. mlo-Based Resistance
mlo-based resistance, initially detected as a natural mutation in an Ethiopian barley cultivar, was successfully introduced in Europe in agricultural programs conferring a broad-spectrum resistance against PM in barley [53][54]. Inactivation of barley MLO protein leads to an enhanced hydrogen peroxide accumulation in the epidermal cells and to cell death in the mesophyll, preventing Bgh penetration [55].
Recently, the barley MLO gene has been cloned, and its resistance mechanisms seem to include callose deposition, increased size of plant papilla and cell wall strengthening [56]. Now, more than half of spring barley is largely immune to PM, due to the introgression of mlo resistance into a broad panel of varieties [57]. Furthermore, researchers found that mlo-based resistance is also a feature of the dicotyledonous Arabidopsis thaliana [58] and many other plant species, such as cucumber [59], tobacco [60], apple [61], pea [62][63] and tomato [64]. mlo-based resistance mechanisms are generally different among plant species. In peas, two recessively inherited genes (er1 and er2), representing the major natural sources of resistance against PM, are both responsible for a defense mechanism independent from HR and associated with the early interruption of pathogenesis after the differentiation of fungal appressoria [62][63]. In tomatoes, the loss-of-function of the MLO gene SlMLO1 leads to a particular form of PM resistance, called ol-2, almost completely preventing pathogen penetration through the apposition of papillae at plant–pathogen interaction sites [64]. This resistance is caused by a natural polymorphism, resulting in a small deletion within the MLO coding region.
To date, mlo resistance has been found as a natural mutation in several crops or produced through induced mutagenesis, gene silencing or gene knock-out [29].
Structural and functional analyses of MLO proteins revealed that the conserved calmodulin-binding domain (CaMBD) seems to be required for full susceptibility to PM infection in barley [65].
Moreover, MLO proteins are characterized by four conserved cysteines [66], and novel conserved peptide domains have been discovered [67]. However, little is known about the molecular function and biochemical activity of these proteins.
MLO genes are found in many crop species, including angiosperms, gymnosperms, lycophytes, bryophytes, algae and other unicellular eukaryotes [19], suggesting that MLO is an ancient eukaryotic protein. To date, a total of ~200 MLO genes have been identified, which are characterized by rich nucleotide diversity and only partially containing a CaMBD [68].
MLO genes encode plant-specific proteins sorted in seven conserved clades, according to the most common classification [29], with IV and V clades appearing to be associated with MLO proteins involved in PM susceptibility in monocots and dicots, respectively [29][69].
Although mlo-based resistance genes have been investigated in several monocot and dicot species, they have been poorly studied in cannabis, as well as other genes involved in disease defense mechanisms [9]. However, in recent years, investigations about MLO genes revealed many key features and characteristics of this family in cannabis, such as the presence of seven transmembrane domains, the presence of the MLO functional domain and the presence of all seven clades, similarly to other crops [19].
Furthermore, to date, barley (Hordeum vulgare) mlo genes are the only race-nonspecific BSR mlo genes identified [31], but their effective and durable resistance has encouraged the identification and characterization of many other MLO orthologs in several plant species, such as Arabidopsis AtMLO2, AtMLO6 and AtMLO12 [58] and cucumber CsaMLO8 [59], in addition to the already mentioned tomato SlMLO1 [64] and pea Er1/PsMLO1 PM [62][63].
3. Powdery Mildew Resistance in Cannabis
Cannabis plants are susceptible to the predominant PM pathogen (Golovinomyces spp.) [10][11][70][71]. Symptoms initially appear as white circular patches of ectophytic mycelia and conidia on the cannabis leaf surface, which later cover the entire surface, and then flowers and buds [10].
Golovinomyces species were found to be a strong post-harvest contaminant of cannabis [18]. These species are G. ambrosiae, G. spadiceus and G. cichoracearum [11][72], whose morphological characters overlapped with several Golovinomyces spp. Furthermore, according to a recent Golovinomyces taxonomic revision based on a multi-locus phylogenetic examination, G. ambrosiae and G. spadiceus were found to form a single undifferentiated clade [73].
In spite of the fact that the vast majority of PM infections in cannabis come from Golovinomyces, another fungal species has been showed to infect this crop, the Podosphaera macularis, which commonly targets hop plants [74][75]. Interestingly, a host-resistance response to this species was observed in ‘TJ’s CBD’, a cannabis cultivar susceptible to G. ambrosiae [76]. This suggests that, in this cultivar, an R gene conferring resistance to P. macularis may be found. Symptoms are evident on foliage, but they are mainly localized on inflorescences in the lower portions of the plant [74]. In greenhouse environments, G. ambrosiae was the most common PM pathogen, while P. macularis was found in plants located in the fields [75]. To date, the P. macularis ability to expand to other sites is still not known [75].
In a recent study [19], CsMLO genes were characterized and their role in PM susceptibility as negative regulatory factors in the cannabis immune system was underlined. Here, the analysis was carried out using the genomes of the ‘Purple Kush’ and ‘Finola’ cannabis cultivars [77], of ‘CBDRx’ [78] and of female and male ‘Jamaican Lion’ [79]. The CsMLO genes study revealed particular amino acid positions, which are present in well-conserved regions, and the phylogenetic analysis of fifteen of them showed that, in all the considered genomes, seven distinct clades (I–VII) were present, as reported in other crops. The focus was on two genes of clade V, CsMLO1 and CsMLO4, both associated with seven transmembrane domains. In fact, the expression analysis revealed that they are remarkably up-regulated during G. ambrosiae infection and were identified as candidates potentially involved in PM susceptibility. The study also included the analysis of amino acids within CsMLO1 and CsMLO4 genes in ~30 commercial cannabis cultivars, revealing several variations, which could influence their related protein functions. Furthermore, in the examined genomes, natural loss-of-function mutations in clade V MLOs were not observed, suggesting that a complete resistance to PM could be rare in commercial cannabis cultivars. Therefore, obtaining a resistant phenotype could be challenging, considering the recessive nature and the genetic redundancy of several CsMLO genes [19].
Another very recent study characterized a new source of PM resistance, confirming the crucial role of MLO genes in PM susceptibility in cannabis [13]. Here, the cannabis cultivar ‘FL 58’ was investigated. The choice of this cultivar was due to the fact that it was subjected to controlled PM inoculation for three consecutive years and no significant infection was observed, thus representing a potential source of PM resistance in C. sativa [80]. Furthermore, two populations, coming from the cross of ‘FL 58’ with the PM susceptible cultivar ‘TJ’s CBD’, were used to identify the genetic basis of PM resistance. These populations were genotyped with single nucleotide polymorphisms (SNPs) and a consensus genetic map was generated. Results showed at least five unique and never identified loci contributing to PM resistance/susceptibility variation. The most associated marker on chromosome 1 was located near the ‘FL 58’ CsMLO1 gene, which was identified as the primary candidate S gene to PM, and it was found to be rare in the cannabis pangenome produced by the Michael lab [13]. Further analyses supported the hypothesis that PM resistance is the effect of the insertion identified in the ‘FL 58’ CsMLO1 sequence, leading to irregular mRNA splicing, and resulting in a premature termination codon. Transcripts encoding a premature stop were found to be ~35 to 65 times more abundant than CsMLO1 full-length transcripts. The consequent strong reduction in functional CsMLO1 proteins could justify the resistance observed in ‘FL 58’ and in other homozygous genotypes [13].
Another significant work showed that the first R gene identified in cannabis was represented by a single dominant locus and was able to confer complete resistance to the PM pathogen G. ambrosiae [18]. Here, for PM pathogen identification, sequence data from 5.8S and 28S rDNA and ITS regions 1 and 2 were generated, and the results showed that the isolate shared 100% sequence homology with G. spadiceus/G. ambrosiae pathogens. The experiments carried out in this study, based on several cannabis cultivars, revealed resistant phenotypes, such as those found in the ‘PNW39’ population, where PM colonies are absent. Then, on the basis of the ‘CBDRx’ cannabis genome annotation, and while adopting the linkage mapping approach with ~10,000 SNP markers, ten candidate genes of a single dominant R gene, named PM1, were identified. This gene resulted in co-localization with the SNP markers LH3804, LH31156, and LH17304 on chromosome 2, and, in the area surrounding the LH3804 locus, a region containing NLRs was identified. More specifically, a cluster of putative disease resistance proteins contained N-terminal coiled-coil (CC) and nucleotide-binding arc (NB-ARC) domains, and two genes with LRR characteristics were detected. Three genes, annotated as tetratricopeptide repeat-containing proteins, were also observed. In conclusion, it can be stated that Mihalyov and Garfinkel’s study [18] provides crucial insights for further genetic cannabis PM resistance research, in order to improve its immunity system.
Furthermore, it is known that NLRs are involved in resistance to PM in several other plant species, like Vitis vinifera [81] and Triticum aestivum [82], and NBS proteins have been associated with candidate PM resistance genes in Humulus lupulus [17]. According to these results and Mihalyov and Garfinkel’s findings [18], NLR-based PM resistance may be hypothesized for cannabis.
Thaumatin-like proteins (TLPs), whose antifungal properties are known [83], were found in hops PM (Podospheara macularis) resistance [84]; however, to date, there is no evidence of this in cannabis.
On the basis of the existing literature and the emerging studies about cannabis PM resistance, a schema of the involved mechanisms is illustrated in Figure 1.
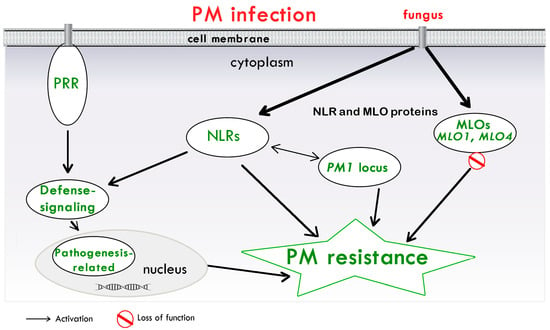
Figure 1. A model of the main mechanisms of PM resistance in cannabis. PAMPs are perceived by membrane-associated PRRs, which activate defense signaling. NLRs recognize pathogen-secreted proteins. These recognitions, in turn, activate immune signaling cascades, resulting in the synthesis of numerous pathogenesis-related proteins to confer PM resistance. Proteins encoded by PM1 gene, represented by a single dominant locus and associated with a region containing NLRs, are shown. Proteins encoded by MLO genes (MLO1 and MLO4), which can lead to PM cannabis resistance, are also included. Abbreviations: Mildew resistance locus o (MLO) gene; NLR, nucleotide-binding and leucine-rich repeat receptor; PM, powdery Mildew; PRR, pattern recognition receptor.
This entry is adapted from the peer-reviewed paper 10.3390/plants13010105
References
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 2019, 3, 430–439.
- Chandran, D.; Wildermuth, M.C. Modulation of Host Endocycle During Plant–Biotroph Interactions. Enzymes 2016, 40, 65–103.
- Hacquard, S. The Genomics of Powdery Mildew Fungi: Past Achievements, Present Status and Future Prospects. Adv. Bot. Res. 2014, 70, 109–142.
- Kuhn, H.; Kwaaitaal, M.; Kusch, S.; Acevedo-Garcia, J.; Wu, H.; Panstruga, R. Biotrophy at its best: Novel findings and unsolved mysteries of the Arabidopsis-powdery mildew pathosystem. Arab. Book 2016, 30, e0184.
- Keinath, A.P.; DuBose, V.B. Controlling powdery mildew on cucurbit rootstock seedlings in the greenhouse with fungicides and biofungicides. Crop Prot. 2012, 42, 338–344.
- Draz, I.S.; Esmail, S.; Abou-Zeid, M.; Essa, T. Powdery mildew susceptibility of spring wheat cultivars as a major constraint on grain yield. Ann. Agric. Sci. 2019, 64, 39–45.
- Clarke, R.C.; Merlin, M.D. Cannabis domestication, breeding history, present day genetic diversity, and future prospects. Crit. Rev. Plant Sci. 2016, 35, 293–327.
- Small, E. Evolution and classification of Cannabis sativa (Marijuana, Hemp) in relation to human utilization. Bot. Rev. 2015, 81, 189–294.
- Sirangelo, T.M.; Ludlow, R.A.; Spadafora, N.D. Molecular Mechanisms Underlying Potential Pathogen Resistance in Cannabis sativa. Plants 2023, 12, 2764.
- Punja, Z.K. Emerging diseases of Cannabis sativa and sustainable management. Pest Manag. Sci. 2021, 77, 3857–3870.
- Wiseman, M.S.; Bates, T.A.; Garfinkel, A.R.; Ocamb, C.M.; Gent, D.H. First report of powdery mildew caused by Golovinomyces ambrosiae on Cannabis sativa in Oregon. Plant Dis. 2021, 106, 2747.
- Dixon, E.; Leonberger, K.; Amsden, B.; Szarka, D.; Munir, M.; Payee, W.; Datnoff, L.; Tubana, B.; Gauthier, N. Suppression of Hemp Powdery Mildew Using Root-Applied Silicon. Plant Health Prog. 2022, 23, 260–264.
- Stack, G.M.; Cala, A.R.; Quade, M.A.; Toth, J.A.; Monserrate, L.A.; Wilkerson, D.G.; Carlson, C.H.; Mamerto, A.; Michael, T.P.; Crawford, S.; et al. Genetic mapping, identification, and characterization of a candidate susceptibility gene for powdery mildew in Cannabis sativa L. Mol. Plant-Microbe Interact. 2023.
- Lyu, D.; Backer, R.G.; Robinson, W.G.; Smith, D.L. Plant-growth promoting rhizobacteria for cannabis production: Yield, cannabinoid profile and disease resistance. Front. Microbiol. 2019, 10, 1761.
- Scott, C.; Punja, Z.K. Evaluation of disease management approaches for powdery mildew on Cannabis sativa L. (marijuana) plants. Can. J. Plant Pathol. 2020, 43, 394–412.
- Henning, G.A.; Gent, D.H.; Townsend, M.S.; Woods, J.L.; Hill, S.T.; Hendrix, D. QTL analysis of resistance to powdery mildew in hop (Humulus lupulus L.). Euphytica 2017, 213, 98.
- Padgitt-Cobb, L.K.; Kingan, S.B.; Henning, J.A. Genomic analysis of powdery mildew resistance in a hop (Humulus lupulus L.) bi-parental population segregating for “R6-locus”. Euphytica 2019, 216, 10.
- Mihalyov, P.D.; Garfinkel, A.R. Discovery and genetic mapping of PM1, a powdery mildew resistance gene in Cannabis sativa L. Front. Agron. 2021, 3, 720215.
- Pépin, N.; Hebert, F.O.; Joly, D.L. Genome-Wide characterization of the MLO gene family in Cannabis sativa reveal two genes as strong candidates for Powdery Mildew susceptibility. Front. Plant Sci. 2021, 12, 729261.
- Cesari, S. Multiple strategies for pathogen perception by plant immune receptors. New Phytol. 2018, 219, 17–24.
- Kourelis, J.; Adachi, H. Activation and regulation of NLR immune receptor networks. Plant Cell Physiol. 2022, 63, 1366–1377.
- Bourras, S.; Kunz, L.; Xue, M.; Praz, C.R.; Müller, M.C.; Kälin, C.; Schläfli, M.; Ackermann, P.; Flückiger, S.; Parlange, F.; et al. The AvrPm3-Pm3 effector-NLR interactions control both race-specific resistance and host-specificity of cereal mildews on wheat. Nat Commun. 2019, 10, 2292.
- Jones, J.D.G.; Vance, R.E.; Dangl, J.L. Intracellular innate immune surveillance devices in plants and animals. Science 2016, 354, aaf6395.
- Marone, D.; Russo, M.A.; Laido, G.; De Leonardis, A.M.; Mastrangelo, A.M. Plant nucleotide binding site-leucine-rich repeat (NBS-LRR) genes: Active guardians in host defense responses. Int. J. Mol. Sci. 2013, 14, 7302–7326.
- Bashir, S.; Rehman, N.; Fakhar Zaman, F.; Naeem, M.K.; Jamal, A.; Tellier, A.; Ilyas, M.; Silva Arias, G.A.; Khan, M.R. Genome-wide characterization of the NLR gene family in tomato (Solanum lycopersicum) and their relatedness to disease resistance. Front. Genet. 2022, 13, 931580.
- Lorek, J.; Panstruga, R.; Hückelhoven, R. The Role of Seven-Transmembrane Domain MLO Proteins, Heterotrimeric G-Proteins, and Monomeric RAC/ROPs in Plant Defense. In Signaling and Communication in Plants Book Series; Baluška, F., Vivanco, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; p. 197.
- Jacott, C.N.; Ridout, C.J.; Murray, J.D. Unmasking Mildew Resistance Locus O. Trends Plant Sci. 2021, 26, 1006–1013.
- Zheng, Z.; Nonomura, T.; Appiano, M.; Pavan, S.; Matsuda, Y.; Toyoda, H.; Wolters, A.M.A.; Visser, R.G.F.; Bai, Y. Loss of function in Mlo orthologs reduces susceptibility of pepper and tomato to powdery mildew disease caused by Leveillula taurica. PLoS ONE 2013, 8, e70723.
- Kusch, S.; Panstruga, R. mlo-Based Resistance: An Apparently Universal “Weapon” to Defeat Powdery Mildew Disease. Mol. Plant-Microbe Interact. 2017, 30, 179–189.
- Brown, J.K.M. Durable resistance of crops to disease: A Darwinian perspective. Annu. Rev. Phytopathol. 2015, 53, 513–539.
- Freisleben, R.; Lein, A. Über die Auffindung einer mehltauresistenten Mutante nach Röntgenbestrahlung einer anfälligen reinen Linie von Sommergerste. Naturwissenschaften 1942, 30, 608.
- Zheng, Z.; Appiano, M.; Pavan, S.; Bracuto, V.; Ricciardi, L.; Visser, R.G.F.; Wolters, A.M.A.; Bai, Y. Genome-wide study of the tomato SlMLO gene family and its functional characterization in response to the powdery mildew fungus Oidium neolycopersici. Front. Plant Sci. 2016, 7, 380.
- Kou, Y.; Wang, S. Broad-spectrum and durability: Understanding of quantitative disease resistance. Curr. Opin. Plant Biol. 2010, 13, 181–185.
- Li, W.; Deng, Y.; Ning, Y.; He, Z.; Wang, G. Exploiting Broad-Spectrum Disease Resistance in Crops: From Molecular Dissection to Breeding. Annu. Rev. Plant Biol. 2020, 71, 575–603.
- Hu, K.; Cao, J.; Zhang, J.; Xia, F.; Ke, Y.; Zhang, H.; Xie, W.; Liu, H.; Cui, Y.; Cao, Y.; et al. Improvement of multiple agronomic traits by a disease resistance gene via cell wall reinforcement. Nat. Plants 2017, 3, 17009.
- Bentham, A.R.; De la Concepcion, J.C.; Mukhi, N.; Zdrzałek, R.R.; Draeger, M.; Gorenkin, D.; Hughes, R.K.; Banfield, M.J. A molecular roadmap to the plant immune system. JBC 2020, 295, 14916–14935.
- Chiang, Y.; Coaker, G. Effector Triggered Immunity: NLR Immune Perception and Downstream Defense Responses. Arab. Book 2015, 13.
- Andolfo, G.; Dohm, J.C.; Himmelbauer, H. Prediction of NB-LRR resistance genes based on full-length sequence homology. Plant J. 2022, 110, 1592–1602.
- Lee, H.A.; Lee, H.Y.; Seo, E.; Lee, J.; Kim, S.B.; Oh, S.; Choi, E.; Choi, E.; Lee, S.E.; Choi, D. Current understandings of plant nonhost resistance. Mol. Plant-Microbe Interact. 2017, 30, 5–15.
- Boutrot, F.; Zipfel, C. Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu. Rev. Phytopathol. 2017, 55, 257–286.
- Xie, S.S.; Duan, C.G. Epigenetic regulation of plant immunity: From chromatin codes to plant disease resistance. aBIOTECH 2023, 4, 124–139.
- Couto, D.; Zipfel, C. Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 2016, 16, 537–552.
- Zurbriggen, M.D.; Carrillo, N.; Hajirezaei, M.R. ROS signaling in the hypersensitive response: When, where and what for? Plant Signal Behav. 2010, 5, 393–396.
- Ng, D.W.; Abeysinghe, J.K.; Kamali, M. Regulating the Regulators: The Control of Transcription Factors in Plant Defense Signaling. Int. J. Mol. Sci. 2018, 19, E3737.
- Jacob, F.; Kracher, B.; Mine, A.; Seyfferth, C.; Blanvillain-Baufume, S.; Parker, J.E.; Tsuda, K.; Schulze-Lefert, P.; Maekawa, T. A dominant-interfering camta3 mutation compromises primary transcriptional outputs mediated by both cell surface and intracellular immune receptors in Arabidopsis thaliana. New Phytol. 2018, 217, 1667–1680.
- Peng, Y.; van Wersch, R.; Zhang, Y. Convergent and Divergent Signaling in PAMP-Triggered Immunity and Effector-Triggered Immunity. Mol. Plant Microbe Interact. 2018, 31, 403–409.
- Tameling, W.I.L.; Vossen, J.H.; Albrecht, M.; Lengauer, T.; Berden, J.A.; Haring, M.A.; Cornelissen, B.J.C.; Takken, F.L.W. Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation. Plant Physiol. 2006, 140, 1233–1245.
- Bonardi, V.; Dangl, J.L. How complex are intracellular immune receptor signaling complexes? Front. Plant Sci. 2012, 3, 237.
- Wu, C.-H.; Abd-El-Haliem, A.; Bozkurt, T.O.; Belhaj, K.; Terauchi, R.; Vossen, J.H.; Kamoun, S. NLR network mediates immunity to diverse plant pathogens. Proc. Natl. Acad. Sci. USA 2017, 114, 8113–8118.
- Bauer, S.; Yu, D.; Lawson, A.W.; Saur, I.M.; Frantzeskakis, L.; Kracher, B.; Logemann, E.; Chai, J.; Maekawa, T.; Schulze-Lefert, P. Theleucine-rich repeats in allelic barley MLA immunereceptors define specificity towards sequence-unrelated powdery mildew avirulence effectors with a predicted common RNase-like fold. PLoS Pathog. 2021, 17, e1009223.
- Narusaka, M.; Shirasu, K.; Noutoshi, Y.; Kubo, Y.; Shiraishi, T.; Iwabuchi, M.; Narusaka, Y. RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant J. 2009, 60, 218–226.
- Schultink, A.; Qi, T.; Lee, A.; Steinbrenner, A.D.; Staskawicz, B. Roq1 mediates recognition of the Xanthomonas and Pseudomonas effector proteins XopQ and HopQ1. Plant J. 2017, 92, 787–795.
- Lyngkjær, M.F.; Østergård, H. Interaction between powdery mildew and barley with mlo5 mildew resistance. Plant Pathol. 1998, 47, 252–258.
- Kang, Y.; Zhou, M.; Merry, A.; Barry, K. Mechanisms of powdery mildew resistance of wheat—A review of molecular breeding. Plant Pathol. 2020, 69, 601–617.
- Piffanelli, P.; Zhou, F.; Casais, C.; Orme, J.; Jarosch, B.; Schaffrath, U.; Collins, N.C.; Panstruga, R.; Schulze-Lefert, P. The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol. 2002, 129, 1076–1085.
- Lyngkjær, M.; Newton, A.; Atzema, J.; Baker, S. The Barley mlo-gene: An important powdery mildew resistance source. Agronomie 2000, 20, 745–756.
- Czembor, J.H.; Czembor, P.C.; Doraczyńska, O.; Pietrusińska, A.; Radecka-Janusik, M. Transfer of the mlo resistance gene into to the genome of winter barley. Prog. Plant Prot. 2016, 56, 379–387.
- Consonni, C.; Humphry, M.E.; Hartmann, H.A.; Livaja, M.; Durner, J.; Westphal, L.; Vogel, J.P.; Lipka, V.; Kemmerling, B.; Schulze-Lefert, P.; et al. Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat. Genet. 2006, 38, 716–720.
- Berg, J.A.; Appiano, M.; Santillán Martínez, M.; Hermans, F.W.K.; Vriezen, W.H.; Visser, R.G.F.; Bai, Y.; Schouten, H.J. A transposable element insertion in the susceptibility gene CsaMLO8 results in hypocotyl resistance to powdery mildew in cucumber. BMC Plant Biol. 2015, 15, 243.
- Fujimura, T.; Sato, S.; Tajima, T.; Arai, M. Powdery mildew resistance in the Japanese domestic tobacco cultivar Kokubu is associated with aberrant splicing of MLO orthologs. Plant Pathol. 2016, 65, 1358–1365.
- Pessina, S.; Pavan, S.; Catalano, D.; Gallotta, A.; Visser, R.G.F.; Bai, Y.; Malnoy, M.; Schouten, H.J. Characterization of the MLO gene family in Rosaceae and gene expression analysis in Malus domestica. BMC Genom. 2014, 15, 618.
- Humphry, M.; Reinstädler, A.; Ivanov, S.; Bisseling, T.; Panstruga, R. Durable broad-spectrum powdery mildew resistance in pea er1 plants is conferred by natural loss-of-function mutations in PsMLO1. Mol. Plant Pathol. 2011, 12, 866–878.
- Pavan, S.; Schiavulli, A.; Appiano, M.; Marcotrigiano, A.R.; Cillo, F.; Visser, R.G.; Bai, Y.; Lotti, C.; Ricciardi, L. Pea powdery mildew er1 resistance is associated to loss-of-function mutations at a MLO homologous locus. Theor. Appl. Genet. 2011, 123, 1425–1431.
- Bai, Y.; Pavan, S.; Zheng, Z.; Zappel, N.F.; Reinstädler, A.; Lotti, C.; De Giovanni, C.; Ricciardi, L.; Lindhout, P.; Visser, R.; et al. Naturally occurring broad-spectrum powdery mildew resistance in a central American tomato accession is caused by loss of Mlo function. Mol. Plant-Microbe Interact. 2008, 21, 30–39.
- Kim, M.C.; Lee, S.H.; Kim, J.K.; Chun, H.J.; Choi, M.S.; Chung, W.S.; Moon, B.C.; Kang, C.H.; Park, C.Y.; Yoo, J.H.; et al. Mlo, a modulator of plant defense and cell death, is a novel calmodulin-binding protein: Isolation and characterization of a rice Mlo homologue. J. Biol. Chem. 2002, 277, 19304–19314.
- Elliott, C.; Müller, J.; Miklis, M.; Bhat, R.A.; Schulze-Lefert, P.; Panstruga, R. Conserved extracellular cysteine residues and cytoplasmic loop-loop interplay are required for functionality of the heptahelical MLO protein. Biochem. J. 2005, 385, 243–254.
- Panstruga, R. Discovery of Novel Conserved Peptide Domains by Ortholog Comparison within Plant Multi-Protein Families. Plant Mol. Biol. 2005, 59, 485–500.
- Shi, J.; Wan, H.; Zai, W.; Xiong, Z.; Wu, W. Phylogenetic Relationship of Plant MLO Genes and Transcriptional Response of MLO Genes to Ralstonia solanacearum in Tomato. Genes 2020, 11, 487.
- Acevedo-Garcia, J.; Kusch, S.; Panstruga, R. Magical mystery tour: MLO proteins in plant immunity and beyond. New Phytol. 2014, 204, 273–281.
- Szarka, D.; Tymon, L.; Amsden, B.; Dixon, E.; Judy, J.; Gauthier, N. First report of powdery mildew caused by Golovinomyces spadiceus on industrial hemp (Cannabis sativa) in Kentucky. Plant Dis. 2019, 103, 1773.
- Maymon, M.; Jerushalmi, S.; Freeman, S. First report of Golovinomyces cichoracearum sensu lato on Cannabis sativa in Israel. New Dis. Rep. 2020, 42, 11.
- Braun, U.; Cook, R.T.A. Taxonomy Manual of the Erysiphales (Powdery Mildews); CBS Biodiversity Series No. 11. CBS-KNAW; Fungal Biodiversity Centre, Ed.: Utrecht, The Netherlands, 2012.
- Qiu, P.-L.; Liu, S.-Y.; Bradshaw, M.; Rooney-Latham, S.; Takamatsu, S.; Bulgakov, T.S.; Tang, S.R.; Feng, J.; Jin, D.N.; Aroge, T.; et al. Multi-locus phylogeny and taxonomy of an unresolved, heterogeneous species complex within the genus Golovinomyces (Ascomycota, Erysiphales), including G. ambrosiae, G. circumfusus and G. spadiceus. BMC Microbiol. 2020, 20, 51.
- Bates, T.; Holzberger-Block, M.; Wiseman, M.; Garfinkel, A.; Gent, D.; Ocamb, C. First report of powdery mildew caused by Podosphaera macularis on hemp in Oregon. Plant Health Prog. 2021, 22, 567–569.
- Punja, Z.K. First report of the powdery mildew pathogen of hops, Podosphaeria macularis, naturally infecting cannabis (Cannabis sativa L., marijuana) plants under field conditions. Can. J. Plant Pathol. 2022, 44, 235–249.
- Weldon, W.A.; Ullrich, M.R.; Smart, L.B.; Smart, C.D.; Gadoury, D.M. Cross-infectivity of powdery mildew isolates originating from hemp (Cannabis sativa) and Japanese hop (Humulus japonicus) in New York. Plant Health Prog. 2020, 21, 47–53.
- Laverty, K.U.; Stout, J.M.; Sullivan, M.J.; Shah, H.; Gill, N.; Holbrook, L.; Deikus, G.; Sebra, R.; Hughes, T.R.; Page, J.E.; et al. A physical and genetic map of Cannabis sativa identifies extensive rearrangements at the THC/CBD acid synthase loci. Genome Res. 2019, 29, 146–156.
- Grassa, C.J.; Weiblen, G.D.; Wenger, J.P.; Dabney, C.; Poplawski, S.G.; Motley, S.T.; Michael, T.P.; Schwartz, C.J. A new Cannabis genome assembly associates elevated cannabidiol (CBD) with hemp introgressed into marijuana. New Phytol. 2021, 230, 1665–1679.
- McKernan, K.J.; Helbert, Y.; Kane, L.T.; Ebling, H.; Zhang, L.; Liu, B.; Eaton, Z.; Sun, L.; Dimalanta, E.; Kingan, S.; et al. Cryptocurrencies and zero mode wave guides: An unclouded path to a more contiguous Cannabis sativa L. genome assembly. OSF 2018.
- Stack, G.M.; Toth, J.A.; Carlson, C.H.; Cala, A.R.; Marrero-González, M.I.; Wilk, R.L.; Gentner, D.R.; Crawford, J.L.; Philippe, G.; Rose, J.K.; et al. Season-long characterization of high-cannabinoid hemp (Cannabis sativa L.) reveals variation in cannabinoid accumulation, flowering time, and disease resistance. Glob. Chang. Biol. Bioenergy 2021, 13, 546–561.
- Goyal, N.; Bhatia, G.; Sharma, S.; Garewal, N.; Upadhyay, A.; Upadhyay, S.K.; Singh, K. Genome-wide characterization revealed role of NBS-LRR genes during powdery mildew infection in Vitis vinifera. Genomics 2020, 112, 312–322.
- He, H.; Zhu, S.; Zhao, R.; Jiang, Z.; Ji, Y.; Ji, J.; Qiu, D.; Li, H.; Bie, T. Pm21, encoding a typical CC-NBS-LRR protein, confers broad spectrum resistance to wheat powdery mildew disease. Mol. Plant. 2018, 11, 879–882.
- Adams, D.J. Fungal cell wall chitinases and glucanases. Microbiology 2004, 150, 2029–2035.
- Kappagantu, M.; Bullock, J.M.; Nelson, M.E.; Eastwell, K.C. Hop stunt viroid: Effect on host (Humulus lupulus) Transcriptome and its interactions with hop Powdery Mildew (Podospheara macularis). Mol. Plant-Microbe Interact. 2017, 30, 842–851.
This entry is offline, you can click here to edit this entry!
