1. Introduction
Wastewater often contains a diverse range of pathogens, including bacteria, viruses, and fungi, due to human and animal waste, industrial discharges, and runoff from contaminated areas [
1]. The existence and abundance of pathogens in wastewater raise significant concerns for public health and the environment. Pathogens in wastewater can cause waterborne diseases, which will produce severe illnesses such as cholera, typhoid, hepatitis, and gastroenteritis [
2]. Moreover, the release of untreated or partially treated wastewater into natural water bodies can contaminate aquatic ecosystems and disrupt the balance of natural environments [
3,
4].
Traditional techniques, such as culture-based methods, microscopic observation, and filtration, have been used to identify and quantify pathogens in wastewater, forming the foundation of early strategies for waterborne pathogen detection [
9]. Culture-based approaches involve the cultivation of microorganisms on specific media, enabling their identification and characterization and contributing to a comprehensive understanding of water quality. The microscopic method focuses on the direct visualization of pathogens under a microscope, offering insights into their morphology and abundance. Filtration techniques concentrate pathogens from high water amounts through filters to capture microorganisms, followed by elution and further analysis.
Meanwhile, molecular methods have been widely utilized for pathogen detection in wastewater. In contrast to culture-based approaches, molecular techniques exhibit higher sensitivity and the capacity to detect low concentrations of target pathogens in wastewater [
10]. Their significantly reduced detection time, often within a few hours, makes them suitable for time-sensitive applications [
10]. Moreover, these methods enable the precise quantification of target DNA, providing valuable insights into pathogen abundance [
11].
The precision and reliability of pathogenic detection in wastewater are challenged by the presence of free nucleic acids, primarily DNA and RNA. These free nucleic acids, originating from both viable and non-viable microorganisms residing in wastewater, introduce complexities that compromise the accuracy of detection and quantification [
12]. To address these challenges, various pretreatment methods have been developed and employed to selectively remove or reduce the levels of free nucleic acids in wastewater samples [
13]. These pretreatment methods can mitigate interference from nontarget nucleic acids, thereby enhancing the sensitivity of pathogen detection techniques and enabling the identification of low pathogen concentrations.
2. Overview of Pretreatment Techniques
Pretreatment methods are designed to selectively remove or reduce the levels of free nucleic acids in wastewater samples. By minimizing the existence of interfering genetic material, pretreatment methods amplify the sensitivity of pathogenic detection, enabling the identification of pathogens even at low concentrations within complex wastewater matrices. These techniques promote the specificity of pathogenic detection assays by diminishing background genetic material, thus facilitating the differentiation between closely related microorganisms or specific pathogenic strains [
25]. Effective pretreatment minimizes the risk of cross-contamination in the laboratory to guarantee the precision and reliability of the results. Additionally, pretreatment techniques contribute to the standardization of the pathogenic detection process, making it more consistent across diverse wastewater samples characterized by varying levels of interfering nucleic acids [
26,
27].
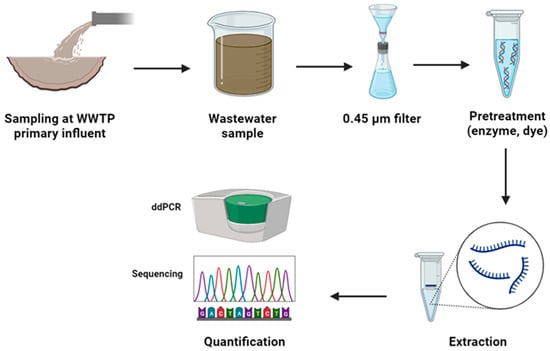
In molecular detection methods, such as PCR, pretreatment is typically conducted during the sample preparation stage (
Figure 1). At this stage, collected wastewater samples may contain a complex mixture of substances, including free nucleic acids, contaminants, and pathogens. Solid particles and debris are separated from the liquid fraction of the sample through filtration. Subsequently, chemical or enzymatic pretreatment methods may be employed to disrupt the structure of cells and pathogens within the sample, thereby releasing genetic material (DNA or RNA) into the solution. Following cell lysis, the sample solution contains a mixture of nucleic acids, including both the genetic material of the target pathogen and interfering free nucleic acids. To selectively isolate the nucleic acids from other components, various extraction methods, such as spin column-based extraction, magnetic bead-based extraction, or organic extraction, have been applied [
28,
29,
30].
Figure 1. Pretreatment methods in the molecular detection methods using PCR techniques.
After pretreatment and extraction, the concentration of the target pathogen’s genetic material can be detected using diverse methods, such as sequencing and real-time PCR. The obtained results are evaluated to detect the presence or absence of the target pathogen. The data derived from the PCR analysis are interpreted, and the quantification of the target pathogen is determined based on the assay’s specific parameters and the standards established during the pretreatment and PCR setup stages.
3. Physical Pretreatment Methods
3.1. Filtration
Filtration, such as microfiltration and ultrafiltration, is a common pretreatment method to remove solid particles, suspended solids, and other particulate matter from wastewater samples. This step plays a crucial role in sample preparation by ensuring that undesired particles or contaminants do not interfere with subsequent molecular analyses like PCR, qPCR, and sequencing [
27]. Moreover, filtration can be effectively utilized to concentrate microorganisms and pathogens, simplifying their detection and analysis within the sample. Notably, filtration efficiently removes contaminants, including organic and inorganic particles, that may hinder molecular detection assays [
31].
Filtration pretreatment encompasses sample preparation, filter setup, sample loading, filtration, recovery, and analyte analysis [
32]. In this context, wastewater samples are initially collected and prepared for filtration. A filtration apparatus, which includes a filter membrane, a filter holder, and a vacuum or pressure source, is assembled. The wastewater sample is then loaded onto the filter membrane, designed to retain particles while allowing the filtrate to pass through. A vacuum or pressure source is employed to force the sample through the filter membrane. Consequently, retained particles, debris, or microorganisms are collected on the filter, while the clarified filtrate is separately obtained. The retained particles or concentrated analytes may be recovered from the filter membrane through techniques like scraping, elution, and dissolution. The clarified filtrate or recovered analytes are ready for molecular detection methods.
3.2. Centrifugation
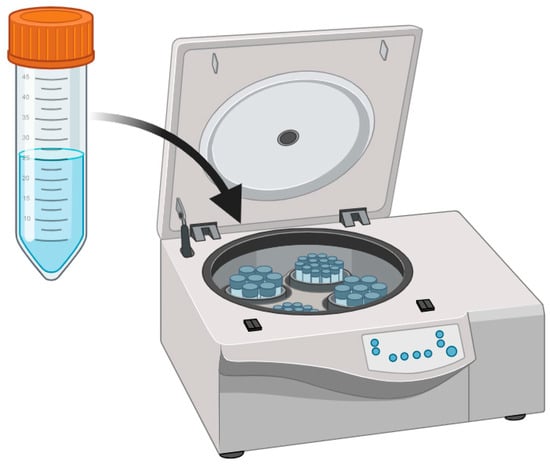
Centrifugation involves using centrifugal force to separate particles, cells, or analytes from the liquid phase within a sample (
Figure 2). It serves the purpose of concentrating cells, microorganisms, and pathogens, simplifying their detection and subsequent analysis in molecular assays. Centrifugation is a key step in many nucleic acid isolation protocols, facilitating the separation of DNA or RNA from other components within the sample [
33].
Figure 2. Centrifugation pretreatment technique.
There are two main types of centrifugation for pretreatment in pathogenic detection. Differential centrifugation involves a sequence of centrifugation steps at varying speeds to separate particles and components of different sizes and densities. This method is frequently utilized to isolate cells, microorganisms, and other analytes from complex samples. In contrast, gradient centrifugation adopts density gradients, typically sucrose or cesium chloride, to separate analytes based on density. It offers a more precise separation and purification of analytes [
34].
The general procedure for centrifugation includes sample preparation, sample loading, centrifugation, supernatant collection, pellet resuspension, and analyte analysis [
35]. Wastewater samples are collected and loaded into centrifuge tubes or containers. The tubes are placed in a centrifuge, where particles and analytes are separated by centrifugal force based on density and size. After centrifugation, the supernatant (liquid phase) is carefully removed, leaving behind a pellet that contains concentrated analytes or particles. The pellet may be resuspended in an appropriate buffer or solvent for further analysis. The concentrated analytes or pelleted materials are ready for molecular detection methods.
3.3. Ultrasonication
Ultrasonication is a valuable pretreatment method for pathogenic detection in molecular analyses like PCR, qPCR, or sequencing. It uses high-frequency sound waves (ultrasound) to disrupt cells, tissues, or particles, thereby facilitating the release of nucleic acids from complex matrices. The mechanism of ultrasonication relies on the formation and rapid collapse of tiny bubbles within a liquid exposed to high-frequency sound waves [
33]. This phenomenon generates localized high-energy forces capable of disrupting cellular membranes, breaking down tissues, and releasing analytes from the sample matrix.
Ultrasonication involves sample preparation, sample loading, ultrasonication, sample cleanup, and analyte recovery. Specifically, wastewater samples are collected and prepared through filtration, dilution, or concentration. The prepared sample is placed in a microcentrifuge or extraction tube, which will be placed in an ultrasonication bath or a probe-type sonicator. High-frequency sound waves are applied to the sample to disrupt cells, tissues, or particles and release analytes. After ultrasonication, the released analytes may undergo additional purification or cleanup steps. The disrupted sample is ready for molecular detection techniques [
36].
4. Chemical Pretreatment Methods
4.1. Precipitation
Precipitation methods can be employed to concentrate and purify DNA or RNA from wastewater samples. They can effectively remove contaminants, impurities, and interfering substances that may affect the quality and accuracy of molecular detection [
33]. It encompasses sample preparation, precipitation reagent addition, mixing and incubation, centrifugation or filtration, supernatant removal, and resuspension [
37]. In particular, wastewater samples are collected, filtered, diluted, or concentrated. An appropriate precipitation reagent, such as alcohol or salt, is added to the wastewater sample to initiate the precipitation process. The sample is mixed to ensure the proper distribution of the precipitation reagent and analyte, and then incubated at specific conditions to allow the analyte to aggregate and precipitate. Centrifugation or filtration will be subsequently employed to separate the precipitate from the liquid phase. The liquid phase (supernatant) is carefully removed, leaving behind the precipitate, which may be resuspended in an appropriate buffer or solvent for further analysis.
4.2. Organic Extraction
Organic extraction can be applied to isolate and purify nucleic acids from wastewater samples. This method involves using organic solvents to selectively separate the nucleic acids from the sample matrix, thereby improving the quality and accuracy of molecular detection [
38]. It comprises sample collection and preparation, organic solvent addition, mixing and phase separation, collection of organic phase, concentration, and resuspension. After the filtration, dilution, or concentration process, an appropriate organic solvent, such as ethanol or isopropanol, is added to the wastewater sample to initiate the extraction process. The sample is mixed to ensure proper contact between the solvent and the analyte. Phase separation, often facilitated by centrifugation or gravity, is then performed to separate the desired analyte from the aqueous matrix. The phase containing the target analyte is carefully collected, while the undesired aqueous phase containing impurities and contaminants is discarded. If the analyte is in low concentrations, the collected phase may undergo concentration steps to increase the analyte’s yield. The isolated and purified analyte is resuspended in an appropriate buffer or solvent for subsequent molecular detection [
39].
4.3. Dye Treatment Method
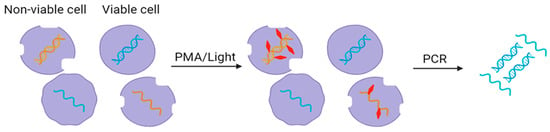
Dye-based pretreatment methods, such as propidium monoazide (PMA), are used to selectively inhibit the detection of non-viable cells or DNA from dead microorganisms in wastewater samples (Figure 3).
Figure 3. Use of PMA dye (red) for removing free nucleic acids in wastewater.
Dyes have the ability to permeate the membranes of dead or non-viable cells, but remain unable to enter the intact membranes of live or viable cells. Once inside the non-viable cell, the dye intercalates with the DNA, forming a covalent bond upon exposure to a light source. This covalent bonding effectively impedes DNA amplification during molecular detection methods like PCR, qPCR, or sequencing [
40]. The outcome is the selective inhibition of DNA amplification from non-viable microorganisms, ensuring that only DNA from vital and intact cells is detected in subsequent analyses. By excluding DNA from non-viable microorganisms, dye-based pretreatment enhances the precision of pathogenic detection in wastewater samples.
In dye-based pretreatment, wastewater samples are collected and prepared for analysis through filtration or concentration. Dye is added to the wastewater sample, permeating the cell membranes of all microorganisms, both non-viable and viable. The sample is incubated in the dark, allowing the dye to penetrate cells and specifically bind to DNA from non-viable cells. After incubation, the sample is exposed to a light source, such as a blue LED, triggering the generation of covalent bonds between the dye and DNA within non-viable cells. DNA extraction is performed on the pretreated sample, and the dye-bound DNA from non-viable cells is purposefully excluded during this extraction. The DNA extracted from viable cells is then subjected to molecular detection methods, offering a highly accurate means of pathogenic detection [
41].
5. Enzymatic Pretreatment Methods
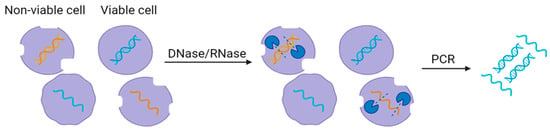
Enzymatic pretreatment methods involve the application of specific enzymes to modify or degrade particular components within the wastewater samples. The selection of the enzyme depends on the analytical objectives, such as the removal of extracellular nucleic acids (e.g., DNase or RNase) (
Figure 4), the degradation of lipids or proteins (e.g., lipases or proteases), or the breakdown of complex structures (e.g., cellulases) [
42].
Figure 4. Use of DNase and RNase enzyme (blue) to remove free nucleic acids in wastewater.
Enzymatic pretreatment includes sample collection, enzyme addition, incubation, enzyme inactivation, sample cleanup, analyte extraction, and molecular detection [
35]. After the filtration process, a specific enzyme is added and mixed with the wastewater sample to ensure uniform distribution. The sample is incubated, permitting the enzyme to act on the targeted components effectively. After incubation, the enzymatic activity is typically paused by heating or adding an enzyme-inactivating reagent. Additional steps for sample cleanup may be performed to eliminate any residual enzymatic activity or reaction byproducts. The treated sample is then subjected to analyte extraction and molecular detection.
This entry is adapted from the peer-reviewed paper 10.3390/applmicrobiol4010001