Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Biochemistry & Molecular Biology
DNA polymerases constitute a versatile group of enzymes that not only perform the essential task of genome duplication but also participate in various genome maintenance pathways, such as base and nucleotide excision repair, non-homologous end-joining, homologous recombination, and translesion synthesis. Polymerases catalyze DNA synthesis via the stepwise addition of deoxynucleoside monophosphates to the 3′ primer end in a partially double-stranded DNA.
- DNA polymerases
- manganese
- translesion synthesis
1. Eukaryotic DNA Polymerases
DNA polymerases can synthesize DNA in a template-dependent manner [1]. They act on a primer/template DNA substrate with a free hydroxyl group at the 3′ position of the sugar moiety of the last nucleotide in the primer. The 3′-OH group is the attachment site during polymerization that proceeds through the sequential addition of dNMPs in an order directed by the template strand. One fundamental cellular task requiring this activity is genome duplication during cell division. Nevertheless, DNA synthesis is needed for several other processes, like the different DNA repair pathways, such as base (BER) and nucleotide excision repair, homologous recombination, non-homologous end-joining (NHEJ), and translesion synthesis (TLS). The structure of the substrate DNA in these pathways varies considerably. Because of this and to fulfill their cellular roles, DNA polymerases became highly specialized [2,3,4]. For example, some DNA polymerases possess exonuclease activity that proofreads mistakes committed by the enzyme. Still, others do not, while some even exhibit 5′- deoxyribophosphate (dRP) lyase, endonuclease, or terminal transferase activities. The fidelity and processivity of polymerases can also differ substantially in accordance with their cellular tasks. The diverse group of eukaryotic DNA polymerases can be classified into four families based on the primary sequence homology of the catalytic domain [5]: the A family contains pols γ, θ, and ν; the B family contains α, δ, ε, and ζ; the Y family includes η, ι, κ, and Rev1; and the X family consists of Polβ, λ, μ, and TdT. Polymerases belonging to the same family share some features, but they all exhibit individual characteristics (Figure 1).
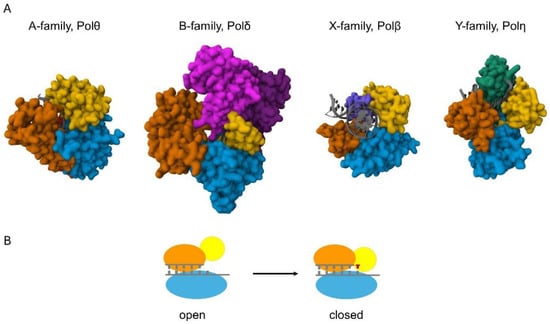
Figure 1. Structures of the catalytic cores of the eukaryotic DNA polymerases. (A) The proteins are shown in surface representation and the DNA helices are shown in cartoon representation (colored grey). The view in all the structures is down the DNA helix axis, except for Polβ, which introduces a 90° bend into the DNA. A family Pols, represented by human Polθ (4X0P), possess palm (blue), fingers (yellow), thumb (orange), and exonuclease (red) domains. Exonuclease domain is positioned behind the palm and thumb domains in the figure. B family Pols, represented by yeast Polδ (3IAY), have N-terminal (purple), exonuclease (magenta), palm (blue), fingers (yellow), and thumb (orange) domains. X family pols, such as human Polβ (4KLE), employ palm, fingers, and thumb domains, as well as a 5′-dRP lyase domain (violet). Y family pols, represented by human Polη (3MR2), have palm, fingers, and thumb domains, and possess a unique polymerase-associated domain (PAD) (green). (B) Schematic of the conformational change of high-fidelity polymerases. The pol binds the DNA in an “open” conformation; then, upon binding the incoming nucleotide, the finger domain (yellow) moves to a “closed” conformation that ensures correct base pairing.
1.1. B Family
The B family members are multisubunit enzymes [11]. This group includes the main replicative polymerases Polε and Polδ. Their role is to carry out faithful duplication of the genomic DNA so that the inheriting material can be transferred to the next generation of cells unchanged. In accordance with this, they are the highest-fidelity DNA polymerases. The high fidelity is attributable to their active centers that impose strict geometric selection during synthesis so that the polymerases cannot accommodate modified, damaged bases and non-Watson–Crick base pairs. In addition, pols δ and ε exhibit a 3′–5′ exonuclease activity that removes accidental errors made by the polymerases, further lowering the error rate during synthesis. Polα is a primase that provides the primer for pols δ and ε, as it can start synthesis de novo on a template strand synthesizing a short 12–15 nt RNA primer by its primase subunit, which is extended with dNTPs by its polymerase subunit. Polα does not have exonuclease activity, and because of this, its fidelity is lowered. Polζ also lacks exonuclease activity. It stands out from the group because it does not work during normal replication. It comes into play when a mismatched or damaged base introduced by other polymerases must be extended. Because its extension activity is essential for synthesis across DNA lesions, Polζ is considered a translesion synthesis polymerase, together with the Y family polymerases.
1.2. Y Family
Polymerases in the Y family are TLS polymerases [12]. They exhibit low fidelity on undamaged DNA due to their spacious, non-selective active center that can accommodate modified, damaged, or mismatched base pairs, in sharp contrast to replicative polymerases. Furthermore, they lack proofreading exonuclease activity. Surprisingly, they can support faithful synthesis across DNA lesions called their cognate lesions, whereas they perform error-prone synthesis across many others. These error-prone polymerases are activated when replication stalls at a DNA lesion site where a TLS polymerase replaces the replicative polymerase to carry out a lesion bypass. Beyond lesion bypass, TLS polymerases must be strictly regulated to avoid the accumulation of excess mutations in the genome. The distributive nature of their synthetic activity supports this confinement. While Pols η and ι can insert nucleotidesacross various DNA lesions, in most cases, they cannot continue the synthesis beyond the inserted nucleotide. Meanwhile, Polκ can work as an inserter and an extender during TLS due to its ability to extend from damaged or mispaired primer ends. Rev1 is very limited as a polymerase: it can catalyze the efficient incorporation of cytosine in a templated manner, whereas it is highly inefficient at inserting other nucleotides. However, it plays an essential scaffolding role during TLS by binding other TLS polymerases.
1.3. X Family
X family polymerases are small, gap-filling repair polymerases that function in the repair of short single-stranded DNA gaps during base excision repair, and in the direct joining of broken DNA ends with minimal or no homology during NHEJ and V(D)J recombination, which is a process that ensures immunoglobulin diversity [13]. In addition to the repair function, pols β, λ, and µ exhibit DNA lesion bypass activity. These polymerases do not have exonuclease activity, but Polβ and λ exhibit a dRP lyase activity that can remove 5′-deoxyribophosphate moieties generated by apurinic/apyrimidinic (AP) endonucleases during BER. Moreover, Polλ and µ were shown to exhibit terminal deoxynucleotidyl transferase activity, like TdT. While Polβ and λ show moderate fidelity, the error rate of Polμ is high, and TdT works primarily in a non-templated fashion and is only expressed in cells engaged in V(D)J recombination [14].
1.4. A Family
The A family member Polγ is responsible for the faithful duplication of the mitochondrial genome, which is supported by its high fidelity and a 3′–5′ proofreading exonuclease activity. In addition, Polγ also has a 5′-dRP lyase activity that is important for its mitochondrial repair function and it shows limited TLS capacity. In contrast, Polθ and ν are low-fidelity enzymes that take part in translesion synthesis, microhomology-mediated end joining, and DNA cross-link repair [15]. Intriguingly, Polθ shows lyase activity and it contains a helicase domain, though helicase activity has not been detected for this protein. Polν is remarkably able to bypass bulky major groove DNA adducts.
2. Metal Ions in DNA Polymerization
All DNA polymerases catalyze the same chemical reaction for which they apply a very similar structural arrangement of the catalytic subunit resembling a human right hand. High-fidelity polymerases undergo a conformational change during catalysis when the right-hand structure transitions from an open to a closed state (Figure 1B) [16]. This conformational change contributes to the fidelity of DNA synthesis. Keeping the analogy, the catalytic subunit has palm, thumb, and finger domains (Figure 1A). The thumb domain binds the double-stranded DNA, the fingers capture the incoming dNTP and the single-stranded template strand, and the palm contains the amino acids that coordinate two divalent metal cations essential for the chemical reaction [17]. The two metals have distinct roles and occupy different positions in the active center [18]. One serves as the catalytic metal at the so-called A site, and the other is the nucleotide metal at the B site. The A site metal helps to lower the pKa of the 3′-OH proton at the primer terminus for nucleophilic attack on the α-phosphate of the incoming nucleotide. The B site metal coordinates the non-bridging oxygens of the triphosphate of the bound nucleotide and helps to neutralize the negative charge during the transition state. After a phosphodiester bond is newly formed between the 3′-O of the primer and the α-phosphate of the dNTP, a pyrophosphate is released. Several structural studies suggest the presence of a third metal during the reaction, but the exact role of the third metal is still debated [19,20]. The identity of the metal cofactor utilized by a given enzyme in the cell is usually uncertain due to technical challenges. Mg2+ has been considered the physiologically relevant activating metal for DNA polymerases because it is abundant in the cell and activates all known DNA polymerases in vitro. Early studies revealed that other metal ions can activate polymerases as well, but usually with much less efficiency and/or fidelity than Mg2+. Moreover, the cellular concentrations of other bivalent metals are significantly lower compared with Mg2+, supporting the pivotal role of Mg2+ in DNA synthesis. Particularly, the intracellular Mn2+ concentration is estimated to be in the µM range (up to 75 µM), whereas that of Mg2+ spans over to the mM range (0.2–7 mM), though the concentration is dependent on the cell type, developmental stage, and organism [21,22,23].
Mutagenic Effect of Manganese
Like Mg2+, Mn2+ can activate all DNA polymerases. However, Cd2+, Co2+, Ni2+, Cr2+, and Mn2+ have been classified as mutagens and potential carcinogens because they cause polymerases to make frequent errors during DNA synthesis in vitro [24,25,26]. Mn2+ was shown to decrease the fidelity of viral, bacterial, and eukaryotic DNA polymerases. In the case of avian myeloblastosis virus DNA polymerase, the efficiency of DNA synthesis was half, and the misincorporation rate was twice or three times higher with Mn2+ compared with Mg2+, depending on the applied Mn concentration. Bacteriophage T4 DNA polymerase and Escherichia coli DNA polymerase I not only misincorporated with elevated rates but removed correctly paired nucleotides with higher rates than mispaired nucleotides if the reactions contained Mn2+ instead of Mg2+ [27]. The misinsertion capability of human DNA Polα was enhanced by Mn2+ as well.
This entry is adapted from the peer-reviewed paper 10.3390/ijms25010363
This entry is offline, you can click here to edit this entry!
