Amino acids are the building blocks of proteins and essential players in pathways such as the citric acid and urea cycle, purine and pyrimidine biosynthesis, and redox cell signaling. Therefore, it is unsurprising that these molecules have a significant role in cancer metabolism and its metabolic plasticity.
- colorectal cancer
- biomarkers
- amino acids
- metabolomics
1. Introduction
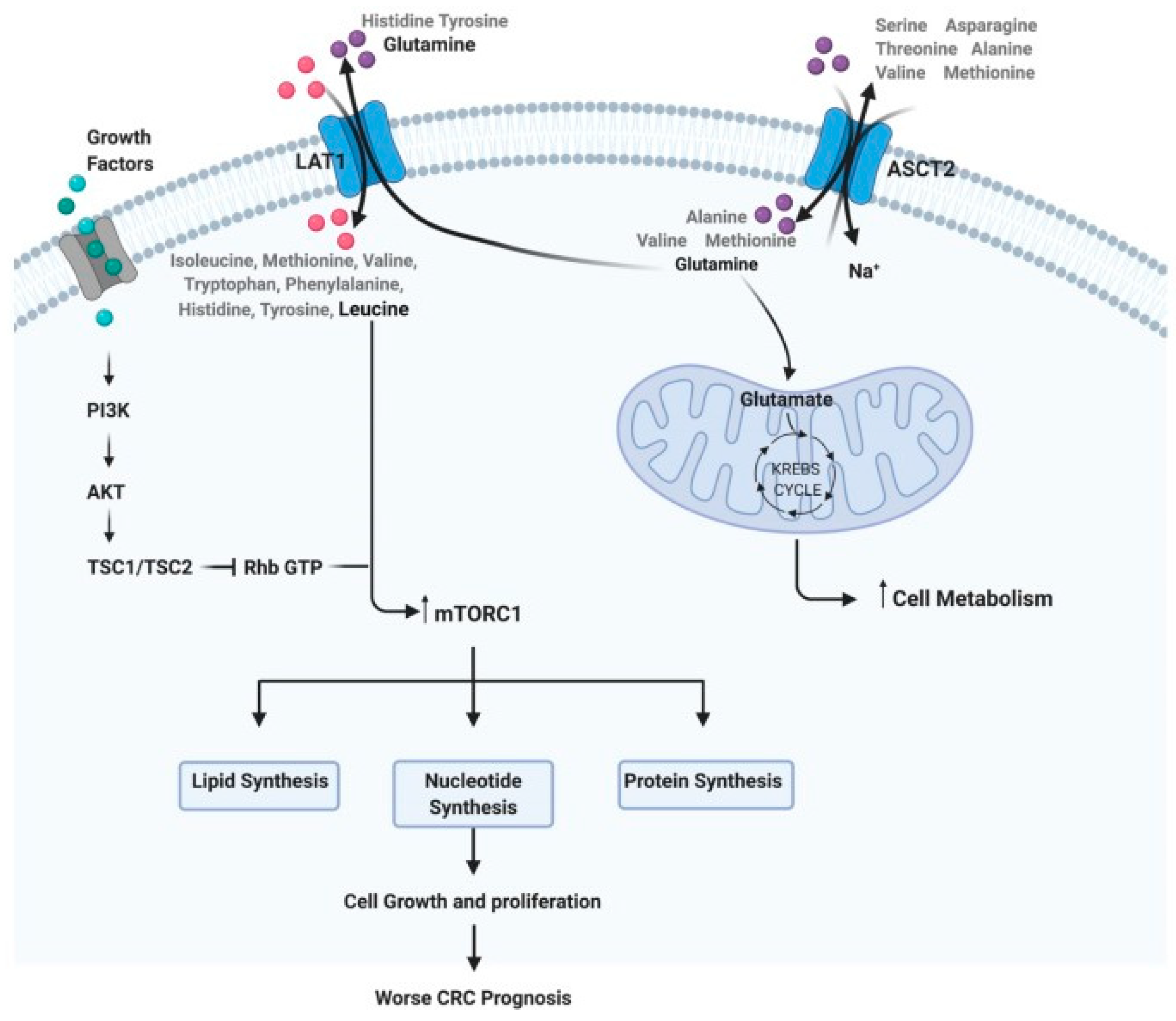
2. Amino Acid Profiling in Colorectal Cancer Patients
2.1. Cancer Tissue Metabolomics
2.2. Serum and Plasma Metabolomics in Colorectal Cancer Patients
2.2.1. Glutamine and Glutamate Metabolism
2.2.2. Glycine and Serine Metabolism
2.2.3. Tryptophan Metabolism
2.2.4. Branched-Chain Amino Acid Metabolism
2.2.5. Proline Metabolism
| Reference | Year | Type of Cancer | TNM Stage | Patients (n) | Healthy Controls (n) | Biological Sample | Laboratory Techniques | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Tan et al. [6] | 2013 | CRC | I/II (68%) | 101 | 102 | Serum | GC-TOFMS/UPLC-QTOFMS | |||
| Chan et al. [15] | 2009 | CRC | Various stages | 31 | 31 | Serum | HR-MAS NMR/GC-MS | |||
| Barberini et al. [24] | 2019 | CRC | Various stages | 15 | 9 | Plasma | GC-MS | |||
| Leichtle et al. [25] | 2012 | CRC | Various stages | 59 | 58 | Serum | ET-MS | |||
| Troisi et al. [29] | 2022 | Adenomas, CRC | I/II | 150 | 50 | Serum | GC-MS | |||
| Gu et al. [30] | 2019 | Adenomas, CRC | Various stages | 62 | 38 | Serum | GC-MS | |||
| Zhu et al. [32] | 2014 | Adenomas, CRC | Various stages | 142 | 92 | Serum | LC-TMS | |||
| Nishiumi et al. [37] | 2017 | Adenomas, CRC | I/II | 282 | 291 | Plasma | GC-T/QMS | |||
| Nishiumi et al. [38] | 2012 | CRC | I/II | 59 | 63 | Serum | GC-MS | |||
| Geijsen et al. [40] | 2019 | CRC | Various stages | 268 | 353 | Plasma | UHPLC-QTOF-MS | |||
| Farshidfar et al. [41] | 2016 | Adenomas, CRC | Various stages | 320 | 254 | Serum | GC-MS | |||
| Ma et al. [42] | 2012 | CRC | Various stages | 30 | 20 | Serum | GC-MS | |||
| Wang et al. [43] | 2017 | CRC | I/II | 55 | 40 | Urine | H-NMR | |||
| Reference | Glutamate | Glutamate | Glycine | Serine | Threonine | Tryptophane | Proline | Valine | Leucine | Isoleucine |
| Tan et al. [6] | D | D | I | D | ||||||
| Chan et al. [15] | I | |||||||||
| Barberini et al. [24] | D | D | D | |||||||
| Leichtle et al. [25] | I | D | ||||||||
| Troisi et al. [29] | D | |||||||||
| Gu et al. [30] | D | I | D | D | D | D | D | D | ||
| Zhu et al. [32] | D | I | D | I | ||||||
| Nishiumi et al. [37] | D | |||||||||
| Nishiumi et al. [38] | D | I | ||||||||
| Geijsen et al. [40] | D | D | ||||||||
| Farshidfar et al. [41] | D | D | D | |||||||
| Ma et al. [42] | D | D | D | |||||||
| Wang et al. [43] | D | D | D | D | ||||||
Legend: I—increase; D—decrease; GC-MS—gas chromatography–mass spectrometry; UHPLC-QTOF-MSa—ultrahigh performance liquid chromatography–quadrupole time-of-flight mass spectrometry; H-NMR—proton nuclear magnetic resonance spectroscopy; ET-MS—electrospray tandem–mass spectrometry; HR-MAS NMR—high-resolution magic angle spinning nuclear magnetic resonance; GC-TOFMS—gas chromatography–time-of-flight mass spectrometry; UPLC-QTOFMS—ultraperformance liquid chromatography–quadrupole time-of-flight mass spectrometry; GC-T/QMS—gas chromatography/triple–quadrupole mass spectrometry.
This entry is adapted from the peer-reviewed paper 10.3390/cancers16010069
References
- Weitz, J.; Koch, M.; Debus, J.; Hohler, T.; Galle, P.R.; Buchler, M.W. Colorectal cancer. Lancet 2005, 365, 153–165.
- Haggar, F.A.; Boushey, R.P. Colorectal cancer epidemiology: Incidence, mortality, survival, and risk factors. Clin. Colon Rectal Surg. 2009, 22, 191–197.
- Strimbu, K.; Tavel, J.A. What are biomarkers? Curr. Opin. HIV AIDS 2010, 5, 463–466.
- Gonzalez-Pons, M.; Cruz-Correa, M. Colorectal Cancer Biomarkers: Where Are We Now? BioMed Res. Int. 2015, 2015, 149014.
- Bedin, C.; Crotti, S.; D’Angelo, E.; D’Aronco, S.; Pucciarelli, S.; Agostini, M. Circulating Biomarkers for Response Prediction of Rectal Cancer to Neoadjuvant Chemoradiotherapy. Curr. Med. Chem. 2020, 27, 4274–4294.
- Tan, B.; Qiu, Y.; Zou, X.; Chen, T.; Xie, G.; Cheng, Y.; Dong, T.; Zhao, L.; Feng, B.; Hu, X.; et al. Metabonomics identifies serum metabolite markers of colorectal cancer. J. Proteome Res. 2013, 12, 3000–3009.
- Agostini, M.; Pucciarelli, S.; Enzo, M.V.; Del Bianco, P.; Briarava, M.; Bedin, C.; Maretto, I.; Friso, M.L.; Lonardi, S.; Mescoli, C.; et al. Circulating cell-free DNA: A promising marker of pathologic tumor response in rectal cancer patients receiving preoperative chemoradiotherapy. Ann. Surg. Oncol. 2011, 18, 2461–2468.
- Brown, R.E.; Short, S.P.; Williams, C.S. Colorectal Cancer and Metabolism. Curr. Color. Cancer Rep. 2018, 14, 226–241.
- Lieu, E.L.; Nguyen, T.; Rhyne, S.; Kim, J. Amino acids in cancer. Exp. Mol. Med. 2020, 52, 15–30.
- Gerner, E.W.; Meyskens, F.L., Jr. Polyamines and cancer: Old molecules, new understanding. Nat. Rev. Cancer 2004, 4, 781–792.
- Dias, F.; Almeida, C.; Teixeira, A.L.; Morais, M.; Medeiros, R. LAT1 and ASCT2 Related microRNAs as Potential New Therapeutic Agents against Colorectal Cancer Progression. Biomedicines 2021, 9, 195.
- Hirayama, A.; Kami, K.; Sugimoto, M.; Sugawara, M.; Toki, N.; Onozuka, H.; Kinoshita, T.; Saito, N.; Ochiai, A.; Tomita, M.; et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 2009, 69, 4918–4925.
- Gao, P.; Zhou, C.; Zhao, L.; Zhang, G.; Zhang, Y. Tissue amino acid profile could be used to differentiate advanced adenoma from colorectal cancer. J. Pharm. Biomed. Anal. 2016, 118, 349–355.
- Denkert, C.; Budczies, J.; Weichert, W.; Wohlgemuth, G.; Scholz, M.; Kind, T.; Niesporek, S.; Noske, A.; Buckendahl, A.; Dietel, M.; et al. Metabolite profiling of human colon carcinoma--deregulation of TCA cycle and amino acid turnover. Mol. Cancer 2008, 7, 72.
- Chan, E.C.; Koh, P.K.; Mal, M.; Cheah, P.Y.; Eu, K.W.; Backshall, A.; Cavill, R.; Nicholson, J.K.; Keun, H.C. Metabolic profiling of human colorectal cancer using high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy and gas chromatography mass spectrometry (GC/MS). J. Proteome Res. 2009, 8, 352–361.
- Chang, L.C.; Fan, C.W.; Tseng, W.K.; Chen, J.R.; Chein, H.P.; Hwang, C.C.; Hua, C.C. Immunohistochemical Study of the Nrf2 Pathway in Colorectal Cancer: Nrf2 Expression is Closely Correlated to Keap1 in the Tumor and Bach1 in the Normal Tissue. Appl. Immunohistochem. Mol. Morphol. AIMM Off. Publ. Soc. Appl. Immunohistochem. 2013, 21, 511–517.
- Piotto, M.; Moussallieh, F.-M.; Dillmann, B.; Imperiale, A.; Neuville, A.; Brigand, C.; Bellocq, J.-P.; Elbayed, K.; Namer, I. Metabolic characterization of primary human colorectal cancers using high resolution magic angle spinning 1 h magnetic resonance spectroscopy. Metabolomics 2009, 5, 292–301.
- Yang, Y.; Zhang, F.; Gao, S.; Wang, Z.; Li, M.; Wei, H.; Zhong, R.; Chen, W. Simultaneous Determination of 34 Amino Acids in Tumor Tissues from Colorectal Cancer Patients Based on the Targeted UHPLC-MS/MS Method. J. Anal. Methods Chem. 2020, 2020, 4641709.
- Qiu, Y.; Cai, G.; Zhou, B.; Li, D.; Zhao, A.; Xie, G.; Li, H.; Cai, S.; Xie, D.; Huang, C.; et al. A distinct metabolic signature of human colorectal cancer with prognostic potential. Clin. Cancer Res. An. Off. J. Am. Assoc. Cancer Res. 2014, 20, 2136–2146.
- Ong, E.S.; Zou, L.; Li, S.; Cheah, P.Y.; Eu, K.W.; Ong, C.N. Metabolic profiling in colorectal cancer reveals signature metabolic shifts during tumorigenesis. Mol. Cell. Proteom. MCP 2010.
- Okada, A.; Takehara, H.; Yoshida, K.; Nishi, M.; Miyake, H.; Kita, Y.; Komi, N. Increased aspartate and glutamate levels in both gastric and colon cancer tissues. Tokushima J. Exp. Med. 1993, 40, 19–25.
- Lin, Y.; Ma, C.; Bezabeh, T.; Wang, Z.; Liang, J.; Huang, Y.; Zhao, J.; Liu, X.; Ye, W.; Tang, W.; et al. (1) H NMR-based metabolomics reveal overlapping discriminatory metabolites and metabolic pathway disturbances between colorectal tumor tissues and fecal samples. Int. J. Cancer. 2019, 145, 1679–1689.
- Rodriguez-Tomas, E.; Arenas, M.; Gomez, J.; Acosta, J.; Trilla, J.; Lopez, Y.; Arquez, M.; Torres, L.; Araguas, P.; Hernandez-Aguilera, A.; et al. Identification of potential metabolic biomarkers of rectal cancer and of the effect of neoadjuvant radiochemotherapy. PLoS ONE 2021, 16, e0250453.
- Barberini, L.; Restivo, A.; Noto, A.; Deidda, S.; Fattuoni, C.; Fanos, V.; Saba, L.; Zorcolo, L.; Mussap, M. A gas chromatography-mass spectrometry (GC-MS) metabolomic approach in human colorectal cancer (CRC): The emerging role of monosaccharides and amino acids. Ann. Transl. Med. 2019, 7, 727.
- Leichtle, A.B.; Nuoffer, J.M.; Ceglarek, U.; Kase, J.; Conrad, T.; Witzigmann, H.; Thiery, J.; Fiedler, G.M. Serum amino acid profiles and their alterations in colorectal cancer. Metabolomics 2012, 8, 643–653.
- Miyagi, Y.; Higashiyama, M.; Gochi, A.; Akaike, M.; Ishikawa, T.; Miura, T.; Saruki, N.; Bando, E.; Kimura, H.; Imamura, F.; et al. Plasma free amino acid profiling of five types of cancer patients and its application for early detection. PLoS ONE 2011, 6, e24143.
- Lee, J.C.; Chen, M.J.; Chang, C.H.; Tiai, Y.F.; Lin, P.W.; Lai, H.S.; Wang, S.T. Plasma amino acid levels in patients with colorectal cancers and liver cirrhosis with hepatocellular carcinoma. Hepato-Gastroenterol. 2003, 50, 1269–1273.
- Wise, D.R.; Thompson, C.B. Glutamine addiction: A new therapeutic target in cancer. Trends Biochem. Sci. 2010, 35, 427–433.
- Troisi, J.; Tafuro, M.; Lombardi, M.; Scala, G.; Richards, S.M.; Symes, S.J.K.; Ascierto, P.A.; Delrio, P.; Tatangelo, F.; Buonerba, C.; et al. A Metabolomics-Based Screening Proposal for Colorectal Cancer. Metabolites 2022, 12, 110.
- Gu, J.; Xiao, Y.; Shu, D.; Liang, X.; Hu, X.; Xie, Y.; Lin, D.; Li, H. Metabolomics Analysis in Serum from Patients with Colorectal Polyp and Colorectal Cancer by 1H-NMR Spectrometry. Dis. Markers 2019, 2019, 3491852.
- Zhu, J.; Djukovic, D.; Deng, L.; Gu, H.; Himmati, F.; Chiorean, E.G.; Raftery, D. Colorectal cancer detection using targeted serum metabolic profiling. J. Proteome Res. 2014, 13, 4120–4130.
- Qiu, Y.; Cai, G.; Su, M.; Chen, T.; Zheng, X.; Xu, Y.; Ni, Y.; Zhao, A.; Xu, L.X.; Cai, S.; et al. Serum metabolite profiling of human colorectal cancer using GC-TOFMS and UPLC-QTOFMS. J. Proteome Res. 2009, 8, 4844–4850.
- Pompella, A.; Visvikis, A.; Paolicchi, A.; De Tata, V.; Casini, A.F. The changing faces of glutathione, a cellular protagonist. Biochem. Pharmacol. 2003, 66, 1499–1503.
- Uyttenhove, C.; Pilotte, L.; Theate, I.; Stroobant, V.; Colau, D.; Parmentier, N.; Boon, T.; Van den Eynde, B.J. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat. Med. 2003, 9, 1269–1274.
- Huang, A.; Fuchs, D.; Widner, B.; Glover, C.; Henderson, D.C.; Allen-Mersh, T.G. Serum tryptophan decrease correlates with immune activation and impaired quality of life in colorectal cancer. Br. J. Cancer 2002, 86, 1691–1696.
- Wang, X.; Liu, H.; Shi, L.; Yu, X.; Gu, Y.; Sun, X. LINP1 facilitates DNA damage repair through non-homologous end joining (NHEJ) pathway and subsequently decreases the sensitivity of cervical cancer cells to ionizing radiation. Cell Cycle 2018, 17, 439–447.
- Nishiumi, S.; Kobayashi, T.; Ikeda, A.; Yoshie, T.; Kibi, M.; Izumi, Y.; Okuno, T.; Hayashi, N.; Kawano, S.; Takenawa, T.; et al. A novel serum metabolomics-based diagnostic approach for colorectal cancer. PLoS ONE 2012, 7, e40459.
- Nishiumi, S.; Kobayashi, T.; Kawana, S.; Unno, Y.; Sakai, T.; Okamoto, K.; Yamada, Y.; Sudo, K.; Yamaji, T.; Saito, Y.; et al. Investigations in the possibility of early detection of colorectal cancer by gas chromatography/triple-quadrupole mass spectrometry. Oncotarget 2017, 8, 17115–17126.
- Wu, J.; Wu, M.; Wu, Q. Identification of potential metabolite markers for colon cancer and rectal cancer using serum metabolomics. J. Clin. Lab. Anal. 2020, 34, e23333.
- Geijsen, A.; Brezina, S.; Keski-Rahkonen, P.; Baierl, A.; Bachleitner-Hofmann, T.; Bergmann, M.M.; Boehm, J.; Brenner, H.; Chang-Claude, J.; van Duijnhoven, F.J.B.; et al. Plasma metabolites associated with colorectal cancer: A discovery-replication strategy. Int. J. Cancer. 2019, 145, 1221–1231.
- Farshidfar, F.; Weljie, A.M.; Kopciuk, K.A.; Hilsden, R.; McGregor, S.E.; Buie, W.D.; MacLean, A.; Vogel, H.J.; Bathe, O.F. A validated metabolomic signature for colorectal cancer: Exploration of the clinical value of metabolomics. Br. J. Cancer 2016, 115, 848–857.
- Ma, Y.; Zhang, P.; Wang, F.; Liu, W.; Yang, J.; Qin, H. An integrated proteomics and metabolomics approach for defining oncofetal biomarkers in the colorectal cancer. Ann. Surg. 2012, 255, 720–730.
- Wang, Z.; Lin, Y.; Liang, J.; Huang, Y.; Ma, C.; Liu, X.; Yang, J. NMR-based metabolomic techniques identify potential urinary biomarkers for early colorectal cancer detection. Oncotarget 2017, 8, 105819–105831.
