Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Clinical models are simple algorithms, developed by the medical community, that predict the functional outcome usually from no more than ten variables using simple arithmetic, as they are meant to be computed by humans.
- deep learning
- ischemic stroke
- modified Rankin Scale (mRS)
1. Introduction
According to the World Stroke Organization, each year, there are over 7.6 million new ischemic strokes, which corresponds to more than 62% of all strokes. Of these annual ischemic strokes, 3.3 million result in death. Additionally, ischemic stroke patients collectively lose more than 63 million healthy years, due to stroke related death and disabilities, each year [1].
Naturally, both the patient and family want to have early information regarding stroke prognosis. Accurate early prediction of post-stroke disability is crucial to both the patient and family to inform actions that should be taken to adapt to a new reality. Additionally, the administration of the thrombolytic drug usually given to these patients can only be done in a limited time frame, and is not risk free [2]. Therefore, in a future where a post-stroke functional outcome predictor is available, these risk factors could be better considered by physicians, which might also allow the use of personalized treatments [3].
The patient’s functional outcome is commonly considered three months after onset and measured by the modified Rankin Scale (mRS), which is an integer scale that goes from zero to six, where zero corresponds to full independence and six corresponds to death [4][5]. Several models have been proposed by the medical and machine learning (ML) communities to predict this variable. As shown in Figure 1, each approach gets its name from the type of data ingested:
-
Tabular approach: only demographic, health records and stroke characterization variables;
-
Image-only approach: only brain imaging data;
-
Hybrid approach: both tabular variables and brain imaging data.
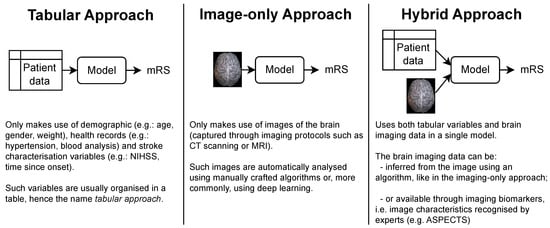
Figure 1. The three main approaches for predicting the mRS. Notably, each approach is characterized only by the type of data it uses. In particular, no assumption is made on how these data are given to the model or how it is processed. This means that the brain imaging data can be the raw brain scans or a series of variables that describe it (for example, image biomarkers).
In terms of the complexity of the models that have been proposed for each approach, the tabular models are by far the simplest. Since their only input is a series of patient variables, they do not require an analysis of the brain scans, which either requires the attention of human experts or the use of sophisticated imaging algorithms, usually involving deep learning.
While simple, the tabular models do not have access to imaging data available in brain scans, such as head CTs. Such scans are collected as part of standard patient care [6], and are known to have relevant information for the prediction of patients’ functional outcome, despite the fact that early admission brain CT scans of ischemic stroke patients only exhibit subtle visual changes [7]. For example, lower ASPECT scores (a score used to systematize the evaluation of the brain damage seen in NCCTs [8]) are correlated with poorer outcomes. Also, the presence and location of vessel occlusions, as well as infarct size, can be estimated from CTA scans and both these variables are correlated with the mRS [7][9]. These findings support the use of the image-only and hybrid approaches (if the brain image scans did not contain predictive information, there would be no point in incorporating them in the models). However, image-only models often underperform relative to the other approaches, and the hybrid models are usually only marginally better than their tabular counterparts.
2. Modified Rankin Scale Prediction
Clinical models are simple algorithms, developed by the medical community, that predict the functional outcome usually from no more than ten variables using simple arithmetic, as they are meant to be computed by humans [3]. The ASTRAL [10], DRAGON [11] and THRIVE [12] scores are examples of such models (following the terminology set in the previous section, ASTRAL [10] is a tabular model, while DRAGON [11] and THRIVE [12] are hybrid models, as they have imaging biomarkers as input).
Machine learning (ML) models are algorithms that can learn from and make predictions on data. They do this by learning the relationships, patterns, and insights contained within the data during the learning (also known as “training”) process. After training, the resulting model can then be used to make prediction on new (unseen) data, a process usually known as “inference”. Because both the training and inference stages are meant to be done by a computer, the ML models often include larger subsets of input variables and also combine them in more complex ways than the clinical models. The term “machine learning” is a broad expression that encompasses both deep learning, which is focused on deep neural networks, as well as “classical” ML models, which are often simpler but also more interpretable (it is easier to understand the resulting trained model). Monteiro et al. [3] trained several classical machine learning models like logistic regressions and random forests on various subsets of tabular variables, and found them to be statistically significantly better than the DRAGON [11] and THRIVE [12] models.
One example of an image-only study is Hilbert et al.’s work [13], where the mRS was predicted from CTA scans. Like all CT scans, these are 3D images of the brain, but the authors transformed these volumes into 2D images by using maximum intensity projection (MIP) [14]. They then used a ResNet [15], adapted with receptive field neural networks (RFNNs) [16], to avoid overfitting, as their prediction model. The MIP technique highlights brain arteries in the axial plane which facilitates the detection of occlusions. Indeed, by analyzing the activation mappings of their network, the authors noticed they tended to focus on these occluded arteries.
Unlike Hilbert’s work [13], which used 2D images as input, most studies process the whole CT scans using 3D convolutional neural networks (CNN). This idea was pioneered by Bacchi et al. [17] with the use of a custom 3D CNN, composed of eight layers, that gathered features from NCCTs scans that were later concatenated with features from clinical and demographic variables to make a prediction. Samak et al. [18] also developed a custom CNN to encode NCCT scans and introduced the use of data augmentations, a more thorough pre-processing, focal loss [19] and attention mechanisms [20]. In another work, Samak et al. developed the feature matching auto-encoder (FeMA) [21], another custom CNN that not only predicted the mRS from admission NCCT scans, but also estimated how these scans would look one week later.
In Brugnara et al. [22] and Ramos et al. [23], statistical tests were employed to compare tabular models with hybrid models. In Brugnara’s work, they tested if adding the acute ischemic volumes and ASPECTS biomarkers would improve an otherwise tabular model. They note that despite both variables being strong independent predictors of the target 90 day mRS, there was no clear advantage in adding either of them (nor both) to the model. In Ramos’s work, they tried adding imaging data by using radiomics features and by using deep learning features from a 3D ResNet 10 encoder [15]. In either case, they concluded that adding imaging features did not improve their models’ performance.
This entry is adapted from the peer-reviewed paper 10.3390/diagnostics13243604
References
- Feigin, V.L.; Brainin, M.; Norrving, B.; Martins, S.; Sacco, R.L.; Hacke, W.; Fisher, M.; Pandian, J.; Lindsay, P. World Stroke Organization (WSO): Global Stroke Fact Sheet 2022. Int. J. Stroke 2022, 17, 18–29.
- Wardlaw, J.M.; Murray, V.; Berge, E.; de Zoppo, G.J. Thrombolysis for acute ischaemic stroke. Cochrane Database Syst. Rev. 2014, 7, CD000213.
- Monteiro, M.; Fonseca, A.C.; Freitas, A.T.; Melo, T.; Francisco, A.P.; Ferro, J.M.; Oliveira, A.L. Using Machine Learning to Improve the Prediction of Functional Outcome in Ischemic Stroke Patients. IEEE/ACM Trans. Comput. Biol. Bioinform. 2018, 15, 1953–1959.
- van Swieten, J.C.; Koudstaal, P.J.; Visser, M.C.; Schouten, H.J.; van Gijn, J. Interobserver agreement for the assessment of handicap in stroke patients. Stroke 1988, 19, 604–607.
- Oliveira, G.; Fonseca, A.C.; Ferro, J.M.; Oliveira, A.L. Potential and limitations of computed tomography images as predictors of the outcome of ischemic stroke events: A review. Front. Stroke 2023, 2, 1242901.
- Hopyan, J.; Ciarallo, A.; Dowlatshahi, D.; Howard, P.; John, V.; Yeung, R.; Zhang, L.; Kim, J.; MacFarlane, G.; Lee, T.-Y.; et al. Certainty of Stroke Diagnosis: Incremental Benefit with CT Perfusion over Noncontrast CT and CT Angiography. Radiology 2010, 255, 142–153.
- de Lucas, E.M.; Sánchez, E.; Gutiérrez, A.; Mandly, A.G.; Ruiz, E.; Flórez, A.F.; Izquierdo, J.; Arnáiz, J.; Piedra, T.; Valle, N.; et al. CT Protocol for Acute Stroke: Tips and Tricks for General Radiologists. RadioGraphics 2008, 28, 1673–1687.
- Barber, P.A.; Demchuk, A.M.; Zhang, J.; Buchan, A.M. Validity and reliability of a quantitative computed tomography score in predicting outcome of hyperacute stroke before thrombolytic therapy. Lancet 2000, 355, 1670–1674.
- Fonseca, A.C.; Ferro, J.M. (Eds.) Precision Medicine in Stroke; Springer: Berlin/Heidelberg, Germany, 2021; ISBN 9783030707613.
- Ntaios, G.; Faouzi, M.; Ferrari, J.; Lang, W.; Vemmos, K.; Michel, P. An integer-based score to predict functional outcome in acute ischemic stroke: The ASTRAL score. Neurology 2012, 78, 1916–1922.
- Kent, T.A. Predicting Outcome of IV Thrombolysis–Treated Ischemic Stroke Patients: The Dragon Score. Neurology 2012, 78, 427–432.
- Flint, A.C.; Cullen, S.P.; Faigeles, B.S.; Rao, V.A. Predicting Long-Term Outcome after Endovascular Stroke Treatment: The Totaled Health Risks in Vascular Events Score. Am. J. Neuroradiol. 2010, 31, 1192–1196.
- Hilbert, A.; Ramos, L.A.; van Os, H.J.A.; Olabarriaga, S.D.; Tolhuisen, M.L.; Wermer, H.J.H.; Barros, R.S.; van der Schaaf, I.; Dippel, D.; Roos, Y.B.W.E.M.; et al. Data-efficient deep learning of radiological image data for outcome prediction after endovascular treatment of patients with acute ischemic stroke. Comput. Biol. Med. 2019, 115, 103516.
- Fishman, E.K.; Ney, D.R.; Heath, D.G.; Corl, F.M.; Horton, K.M.; Johnson, P.T. Volume Rendering versus Maximum Intensity Projection in CT Angiography: What Works Best, When, and Why. RadioGraphics 2006, 26, 905–922.
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 23–30 June 2016; pp. 770–778.
- Jacobsen, J.-H.; Gemert, V.; Lou, Z.; Smeulders, A.W.M. Structured Receptive Fields in CNNs. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 23–30 June 2016; pp. 2610–2619.
- Bacchi, S.; Zerner, T.; Oakden-Rayner, L.; Kleinig, T.; Patel, S.; Jannes, J. Deep Learning in the Prediction of Ischaemic Stroke Thrombolysis Functional Outcomes. Acad. Radiol. 2020, 27, e19–e23.
- Samak, Z.A.; Clatworthy, P.; Mirmehdi, M. Prediction of Thrombectomy Functional Outcomes Using Multimodal Data. In Proceedings of the Medical Image Understanding and Analysis, 24th Annual Conference, MIUA 2020, Oxford, UK, 15–17 July 2020; Volume 1248, pp. 267–279.
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal Loss for Dense Object Detection. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2999–3007.
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-Excitation Networks. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018.
- Samak, Z.A.; Mirmehdi, P.; Clatworthy, M. FeMA: Feature matching auto-encoder for predicting ischaemic stroke evolution and treatment outcome. Comput. Med. Imaging Graph. 2022, 99, 102089.
- Brugnara, G.; Mihalicz, P.; Herweh, C.; Schönenberger, S.; Purrucker, J.; Nagel, S.; Ringleb, P.A.; Bendszus, M.; Möhlenbruch, M.A.; Neuberger, U. Clinical value of automated volumetric quantification of early ischemic tissue changes on non-contrast CT. J. Neurointerv. Surg. 2022, 15, e178–e183.
- Ramos, L.A.; van Os, H.; Hilbert, A.; Olabarriaga, S.D.; Lugt, A.; Roos, Y.B.W.E.; Zwam, W.H.; Walderveen, M.A.A.; Zwinderman, A.H.; Strijkers, G.J.; et al. Combination of Radiological and Clinical Baseline Data for Outcome Prediction of Patients With an Acute Ischemic Stroke. Front. Neurol. 2022, 13, 809343.
This entry is offline, you can click here to edit this entry!
