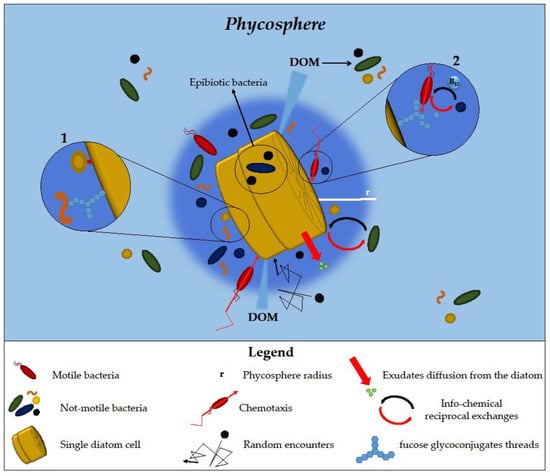
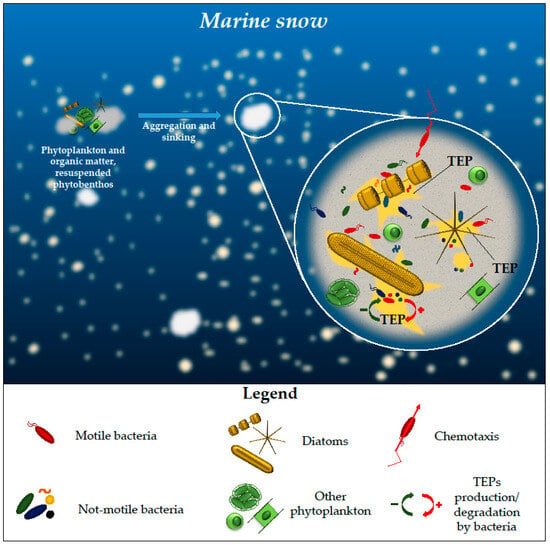
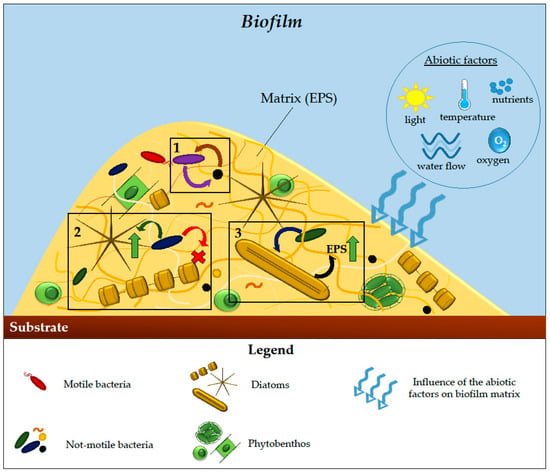
Nutrient availability can also influence bacteria–diatom population dynamics, as demonstrated by the capability of
A. macleodii to have positive, negative, or neutral interactions with the diatom
P. tricornutum, depending on nitrogen availability [
82]. Specifically,
A. macleodii establishes commensal relationships with
P. tricornutum during its exponential growth, with no impact on diatom growth, when nitrogen is abundant, benefiting from the dissolved organic carbon (DOC) released by this diatom species. Later, during the stationary phase, both diatoms and bacteria continue to increase their cell numbers, suggesting the occurrence of a cooperative interaction with the bacteria providing the nitrate to the diatom for their growth, while diatoms provide the organic matter to bacteria. Conversely, the addition of DOC to the media at the diatom’s early growth phase, before nitrate becomes depleted, triggers
A. macleodii proliferation and reduces
P. tricornutum growth, suggesting that in the presence of sufficient DOC, the bacterium competes with the diatom for nitrogen uptake.
3.1. Mutualistic Interactions
A typical mutualistic relationship is based on the exchange of cobalamin (vitamin B
12), for which many diatoms, as well as many other eukaryotic algae, are auxotrophs [
83,
84].
In exchange for organic carbon or nitrogen, selected bacteria and archaea establish symbiosis with diatoms by proving vitamin B
12 that is produced de novo or through modifications of pseudo-cobalamin and other closely related compounds [
97]. However, this system represents an active relationship and not a mere passive exchange of nutrients [
85,
86]. Indeed, to create a specific association with the bacterium
Ruegeria pomeroyi, which supplies diatoms with vitamin B
12,
Thalassiosira pseudonana releases 2,3-dihydroxypropane-1-sulfonate (DHPS). When in co-culture with
T. pseudonana,
R. pomeroyi shows an up-regulation of the genes involved in DHPS catabolism, while the diatom upregulates genes involved in the release of organic compounds, supporting bacterial growth. In a coastal environment in which iron and vitamin B
12 limitations were found, the main vitamin B
12 producer in plankton communities was found to be the Gammaproteobacteria
Oceanospirillaceae ASP10-02a [
87]. This bacterium showed high expression of genes related to organic matter acquisition and cell surface attachment, thus entertaining a mutualistic relationship with phytoplankton, among which diatoms, to fuel vitamin B
12 production. Moreover,
Pseudo-nitzschia subcurvata exuded organic matter that is used by
Sulfitobacter sp. SA1 as a source of nutrients, while the bacterium provides biotin, vitamin B
12, and thiamine that support diatom growth in vitamin-limiting conditions [
88]. SA1 also possesses a catalase that, similarly to what was previously observed in another diatom-
Sulfitobacter association, can further improve
P. subcurvata growth, supporting it in detoxification processes.
Similarly, diatoms unable to fix atmospheric N
2 establish symbiosis with nitrogen-fixing cyanobacteria, giving in return amino acids and organic carbon [
8,
89]. N
2-fixing cyanobacteria can be both obligate and facultative. As an example,
Richelia intracellularis is an obligate symbiont adapted to live inside the
Rhizosolenia and
Hemiaulus genera frustules and is also transmitted to the host’s next generation [
90].
Certain bacteria use monomethyl amine (MMA), ubiquitously present in the ocean, as sources of organic carbon, energy, and nitrogen (or the sole nitrogen source) [
80]. For example, the strain KarMa of the
Rhodobacteraceae Donghicola sp. retrieves nitrogen in the form of ammonium from MMA degradation, providing it to
P. tricornutum, thus sustaining its growth under photoautotrophic conditions. This interaction has a mutualistic character since KarMa growth, in turn, is supported by diatom-released organic carbon. This cross-feeding is widespread, since it was also observed when KarMa was co-cultured with the other two diatoms, i.e.,
Amphora coffeaeformis and
T. pseudonana [
92].
3.2. Facilitative Interactions
The beneficial effect of bacteria on diatom health and growth has been shown in many studies to occur through different mechanisms, always involving metabolite exchange [
67,
98,
99,
100,
101,
102]. Less is known about possible advantages for the bacterial community growing with diatoms, besides the already-ascertained advantage of benefiting from organic matter released by co-occurring diatoms.
Bacteria are able to influence the diatom metabolic profile by stimulating diatom cells towards the synthesis of amino acids and secondary metabolites. In a study conducted by co-culturing
T. pseudonana with the bacterium
Dinoroseobacter shibae, separated by a membrane that allowed only the exchange of chemical signals without any physical contact, diatom abundance was higher in comparison with the axenic algae [
67]. Metabolic activity was also increased, especially with regards to the concentration of some intracellular amino acids and their derivatives, while the general health status did not change.
The bacterium
Bacillus thuringiensis, following sporulation and mother cell lysis, releases compounds among which two diketopiperazines (DKPs), which are able to stimulate the growth of
P. tricornutum as well as its content in neutral lipid [
98].
3.3. Antagonistic Interactions
3.3.1. Inhibitory Effects of Bacteria on Diatoms
Not always the interactions between diatoms and bacteria are beneficial for one or both of them [
4].
Table 3 reports a simplified list of the inhibitory effects exerted by bacteria on diatoms.
Table 3. Examples of antagonistic interactions based on the inhibitory effects of bacteria on diatoms, including the compounds involved. Acronyms: NA = Not Available; OXO12 = N-(3-oxododecanoyl) homoserine lactone; TA12 = OXO12 tetramic acid; HHQ = 2-heptyl-4-quinolone; PHQ = 2-pentyl-4-quinolone; PQ = 2-n-pentyl-4-quinolinol.
| Bacterial Species |
Diatom Species |
Bacterial Compounds |
Effects on Diatoms |
References |
| Croceibacter atlanticus |
Pseudo-nitzschia
multistriata |
NA |
induction of DNA fragmentation |
[21] |
| Methylophaga |
phytoplankton
communities |
NA |
competition for vitamin B12 |
[87] |
| Olleya sp. A30 |
Pseudo-nitzschia
subcurvata |
NA |
growth impairment |
[88] |
| Croceibacter atlanticus |
Thalassiosira
pseudonana |
extracellular metabolites |
inhibition of cell division, alteration of cell morphology, increase in organic matter release |
[104] |
| Maribacter sp. and Marinobacter sp. |
Seminavis robusta |
NA |
negative influence on sexual reproduction rate by affecting
diproline production |
[9,105] |
| marine Proteobacteria |
Phaeodactylum
tricornutum |
OXO12 and TA12 |
inhibition of growth |
[106] |
| Pseudoalteromonas sp. and Alteromonas sp. |
Phaeodactylum
tricornutum |
HHQ |
Growth impairment by inhibition of photosynthetic electron transport and respiration |
[107] |
Thalassiosira weissflogii and Cylindrotheca
fusiformis |
PHQ |
inhibition of growth |
[108] |
Amphora coffeaeformis, Navicula sp., and
Auricula sp. |
PQ |
inhibition of motility |
[109] |
An example is given by the Flavobacter Croceibacter atlanticus, commonly associated with Pseudo-nitzschia multistriata, negatively affecting the growth of this and other diatom genera [21]. It colonises diatom cell surfaces, using their exudates to proliferate until diatoms reach the stationary growth phase. At this point, it leads to diatom death (likely to use the released organic matter), leaving the diatom cell surface before they sink.
Also, vitamin B12-consumer bacteria can establish antagonistic interactions with diatoms, as in the case of Methylophaga, a gammaproteobacteria that competes for vitamin B12 while relying on phytoplankton carbon and energy to sustain its growth [87]. Olleya sp. A30, which possesses genes able to degrade diatom-derived organic compounds, seems be able to enter the frustules, consuming diatom organic matter from the inside [88]. When in co-culture with P. subcurvata, A30 negatively impacts the growth of this diatom by competing for vitamin B12.
Some bacterial genera affect, in a species-specific way, the efficiency of sexual reproduction [105]. Maribacter sp. and Marinobacter sp. cells and their spent medium, for example, negatively affect the sexual reproduction rate of the biofilm-forming pennate diatom S. robusta, generally more efficient in axenicity. On the other hand, Roseovarius sp. increased the proportion of sexually active S. robusta cells.
Some marine proteobacteria frequently associated with diatoms use AHLs as signal molecules in QS [106]. In a biofilm matrix, AHLs can reach high local concentrations and be converted into tetramic acids (TAs). Using synthetic analogues, it was observed that both the N-(3-oxododecanoyl) homoserine lactone (OXO12) and its tetramic acid (TA12) inhibited the growth of the diatom P. tricornutum, with TA12 exerting this action at lower concentrations than OXO12. In addition, increasing concentrations of TA12, able to bind the quinone B (QB) binding site in the photosystem II (PsII), caused a decrease in the diatom’s photosynthetic ability. On the other side, the benthic Nitzschia cf. pellucida seems to be able to disrupt bacterial β-keto-AHL signals using its haloperoxidase system to cleave its halogenated N-acyl chain [110]. Other than AHLs, other QS molecules, i.e., the quinolones, mediate diatom–bacteria interactions [107]. Quinolones are mainly produced by marine bacteria belonging to the Pseudoalteromonas and Alteromonas genera, but also by soil and freshwater bacteria belonging to the Pseudomonas and Burkholderia genera [107].
3.3.2. Algicidal Bacteria
Algicidal bacteria are important components in the determination of phytoplankton successions in marine environments since they can inhibit microalgal growth both through direct contact and the active release of diffusible factors to lyse cells [
35,
111]. The mode of action of algicidal bacteria ranges from high specificity for bacteria that kill only one species to universal for bacteria affecting a broad range of phytoplankton members [
111]. For some bacteria, especially those living in biofilms, the algicidal activity is triggered by the perception of a certain microalgae density through a QS system [
107], while others can switch on and off their algicidal activity depending on nutrient availability, with high nutrient concentrations stimulating the algicidal activity [
112].
The most widespread algicidal bacteria belong to the genera
Saprospira,
Vibrio,
Alteromonas,
Pseudomonas, and
Pseudoalteromonas [
113]. Generally,
Alteromonas,
Pseudoalteromonas, and
Vibrio kill diatoms, releasing diffusible substances, while other genera such as
Cytophaga and
Flavobacterium require direct contact with diatoms [
34]. There are some exceptions reported, and one of them is
Kordia algicida, a
Flavobacterium that uses the QS-controlled release of diffusible proteases to kill a wide range of algal species, including the diatoms
S. costatum,
T. weissflogii [
113],
C. socialis [
111], and
P. tricornutum [
34]. On the contrary,
Chaetoceros didymus is not susceptible to
K. algicida presence, and, surprisingly, the medium in which they are co-cultured is still active against
S. costatum [
34], a species susceptible also to the release of diffusible factors with protease and DNAse activities from other algicidal bacteria [
114].
3.3.3. Inhibitory Effects of Diatoms on Bacterial Growth
Some diatom species are able to activate defence mechanisms to deal with algicidal bacteria (simplified list of examples in
Table 5) [
4].
Table 5. Examples of antagonistic interaction based on diatom inhibition of bacterial growth and of the compounds involved. Acronyms: NA = Not Available; EPA = eicosapentaenoic acid; PA = palmitoleic acid; HTA = (6Z, 9Z, 12Z)-hexadecatrienoic acid.
| Bacterial Species |
Diatom Species |
Diatoms Compounds |
Effects on
Bacteria |
References |
| Kordia algicida |
Chaetoceros didymus |
15-HEPE |
inhibition of growth |
[35] |
| Vibrio alginolyticus, V. campbellii, and V. harveyi |
Nitzschia laevis, two Nitzschia frustulum strains, Navicula incerta, Navicula cf. incerta, and Navicula biskanterae |
NA |
inhibition of growth |
[119] |
| marine and not-marine gram-positive and negative bacteria |
Phaeodactylum tricornutum |
EPA, PA, and HTA |
death |
[120,121,122] |
The inhibitory effects of diatoms on bacterial growth are often mediated by compounds that, in some cases, have been isolated and characterised. PUAs, for example, affect diatom-associated bacterial communities when present in high concentrations [45,123]. Pure 2E,4E-decadienal, 2E,4E-octadienal, and 2E,4E-heptadienal tested on 33 bacterial strains at unnaturally high concentrations showed a variety of effects ranging from concentration-dependent growth inhibition to no effect or growth stimulation [43,123]. However, strain-specific effects on bacterial strains isolated from the Mediterranean Sea were also observed in laboratory experiments using PUA concentrations similar to those found in nature. Indeed, bacteria belonging to Gammaproteobacteria were less affected compared to Rhodobacteraceae, whose abundance decreased by 21%, suggesting a role for PUAs in the regulation of diatom-associated microbiome composition [124].
On the other side, oxylipins (oxygenated PUFA derivatives) are used by
C. didymus against
K. algicida attacks [
34]. Indeed, differently from
S. costatum, which is susceptible to the bacterial attack,
C. didymus uses a wound-dependent defence mechanism to counteract the bacterial attack, which activates the synthesis and release of oxylipins [
35], but also releases proteases [
125]. The most abundant oxylipin released by
C. didymus when attacked by
K. algicida is the hydroxylated eicosapentaenoic acid 15-HEPE, which significantly inhibits bacterial growth [
35].
P. tricornutum cell lysates showed antibacterial activity in vitro, which was attributed to the presence of eicosapentaenoic acid (EPA), palmitoleic acid (PA), and (6Z, 9Z, 12Z)-hexadecatrienoic acid (HTA), active against both marine and non-marine gram-positive and negative bacteria [
120,
121]. The analysis of the concentration of these fatty acids in the different
P. tricornutum morphotypes highlighted higher production by fusiform cells with respect to the oval one [
122].
4. Diatom-Associated Microbiomes
In the last few years, interest in studying the complexity of interactions established in nature between diatoms and their microbiome has increased, thus providing new information on their interplay [
128], the stability of the associations and diatom species specificity [
6], and the seasonality or geographical location influences [
129]. As an example, a recent in situ study performed through metabarcoding of samples collected along the Australian coast in different seasons and locations allowed the characterization of the microbial community and reported that temperature and nutrient composition drive diatom community assemblages in different geographical locations. Moreover, they observed patterns of co-occurrence, conserved across space and time, among certain bacteria belonging to the
Roseobacter and
Flavobacteria clade with the diatoms of the genera
Skeletonema,
Thalassiosira, and
Cylindrotheca [
130], suggesting that species-specific interactions take place between these organisms and that those bacteria may significantly contribute to the seasonal and spatial variability of diatom communities.
In general, the structure of a diatom’s associated microbiomes may be determined, in some cases, by selective processes that are both environmental and host-guided, or in other cases, by neutral lottery-like processes, for which the possibility of a certain bacteria becoming part of a microbiome is proportional to its abundance in the environment [
131].
Chemo-attractant and chemo-repellent compounds, but also secondary metabolites, released by diatoms in the phycosphere [
4,
8,
16], can greatly influence the microbiome composition [
11]. As an example, the analysis of the
P. tricornutum exo-metabolome revealed the production of a pool of metabolites able to shape the associated microbiome composition, among which the 4-hydroxybenzoic acid selectively stimulated the growth of bacteria capable of metabolising and using it as a carbon source [
133].
Axenic
A. glacialis, when re-inoculated with its original microbiome, showed significant changes in its transcriptome and metabolome profiles and started secreting azelaic acid (AA) and rosmarinic acid (RA), able to favour the establishment of selected bacterial species while inhibiting the growth of unnecessary species [
28].
On the other side, some bacteria species are able to control the composition of the microbiome associated with a certain diatom. This is the case of
Phaeobacter inhibens, naturally occurring in association with
T. rotula, which was shown to influence the microbiome assembly associated with the diatom [
128].
Many of the cited studies have been conducted in laboratory conditions and revealed the long-term stability of the associated microbiomes, as for different strains of
Asterionellopsis glacialis and
Nitzschia longissima, which maintained a unique bacterial community for up to one year of growth in the laboratory [
6]. In addition, species belonging to the
Roseobacter clade were constantly present in all the analysed diatom strains and time periods. Also, the microbiomes of different
P. tricornutum strains, despite belonging to different geographical locations, were very similar among each other and remained stable during laboratory cultivation, differing from the ones of other microalgal genera, i.e.,
Entomoneis and
Tetraselmis, cultivated in the same conditions [
134]. However, cultivation parameters conditioned the microbiome composition since, in the same
P. tricornutum strains, it changed slightly when grown in a different medium with a higher nutrient concentration.
5. Most Used Approaches to Study the Bacterial Communities’ Diversity
The technological advancement, especially in omics technologies, is greatly contributing to improving our understanding of microbial interactions, which are the subject of intensive investigations and projects based on the integration of big data coming from different approaches and omics techniques [
139]. This approach integrates the analyses of meta-omics data to predict: (i) potential biotic interactions; (ii) reveal niche spaces; and (iii) guide more focused studies on possible relations and roles of compounds from different organisms composing the bacteria–diatom community. The co-occurrence pattern represents a useful tool to infer possible connections among microorganisms based on their abundances.
Another level of resolution is represented by the “trait-based” approach, which aims to reconstruct community biodiversity by tracking microorganisms’ functional traits, ultimately inferring ecosystem functioning [
141]. In particular, this approach uses functional annotations to cluster genomes by functions that co-evolved and are possibly connected, which allows grouping both known and unknown microorganisms. The use of clustered functions simplifies the analysis of enormous metagenomics datasets and allows the simplification and unravelling of the intricate interconnected fluxes of functions at the base of complex communities [
141,
142].
The bioinformatics pipeline IDBac is a useful tool to integrate information on bacteria, proteins, and secondary metabolite profiles obtained through MALDI-TOF MS analysis [
143]. This approach is based on the assessment of bacterial traits, the identification of phylogenetic relationships, and the comparison of metabolic differences among hundreds of clones in a short time.
At a lower scale, a community made up of a single diatom species and its associated bacteria can be studied by a combination of cell sorting techniques and metagenome sequencing of all the species composing the microbial community. Sequence clustering allows them to associate a set of genomic sequences with each species composing the consortium. Subsequent phylogenetic analysis can lead to identification at a species or genus level [
103,
144]. More specifically, this pipeline includes the utilisation of the NCBI database BLASwares by the Geneious
® software [
144] to identify and edit the 16S rDNA sequences found in the previous step and then the SILVA INcremental Aligner tool (SINA) to align the sequences [
145]. ARB is also used to identify ribosomal sequences not identified with the other tools and steps [
146]. Mothur [
147] and NodeXL are useful tools to cluster the sequences based on their OTU and are used for network analysis and to visualise how communities are interconnected.
6. Potential Biotechnological Applications of Diatom–Bacteria Consortia
Bacteria have been considered for a long time as undesired contaminants in microalgae cultures grown for biotechnological purposes [
154]. This view has now changed, and interactions between microalgae and their microbiome are being investigated for possible useful applications [
155]. Nevertheless, most of the studies produced until now concern the use of consortia made of microalgal species in the
Chlorella,
Scenedesmus,
Tetraselmis, and
Chlamydomonas genera and their associated bacteria for the removal of nutrients from eutrophic waters, wastewater treatment, biofuel, and valuable compound production [
154,
156,
157].
The beneficial cooperative interactions between diatoms and bacteria may provide useful tools to optimise the productivity of microalgae cultivation, since in most cases they culminate in an increase in diatom growth and consequently in their biomass [
68], as for
T. pseudonana, which increased cell density by 35% when cultivated in the presence of
D. shibae [
67]. The stimulatory effect of bacteria on diatom growth finds application mainly in wastewater treatment, bioremediation [
156,
162], and biofuel production [
154]. In this regard, there is an increasing number of studies driving the development of artificial consortia specific to each application based on the interaction mechanisms occurring in the consortia [
157].
The presence of bacteria may positively influence the tolerance and performance of diatoms cultivated in presence of pollutants [
158]. Indeed, it has been observed that the co-occurrence of free-living bacteria belonging to the hydrocarbon-degrading genera
Marivita,
Erythrobacter, and
Alcaligenes with the diatom
Nitzschia sp. isolated from polycyclic aromatic hydrocarbons (PAHs)-contaminated sediments increased the growth rates and enhanced the tolerance of this microalga to PAHs compared to the performances of axenic diatom cultures, finally resulting in increased bioremediation of the PAHs fluoranthene (Flt) and benzo(a)pyrene (BaP) contaminated sites. This improvement in diatom tolerance to contaminants due to their associated bacteria has also been observed for
Thalassiosira delicatula grown in the presence of metals and pesticide mixtures [
163].
During the final steps of wastewater treatment, as well as in other industrial applications such as biofuel production, bacteria that increase microalgae flocculation through polysaccharides or protein production represents a sustainable, efficient, and cost-effective alternative to some toxic flocculation agents [
155,
162]. Nevertheless, the use of bacteria as flocculating agents in industrial applications is poorly explored for diatoms, but rather for other microalgae [
164].
Biofilms dominated by diatoms and bacteria can be also applied in aquaculture to improve larval fish growth [
161]. Indeed, the biofilm forming naturally in
Seriola lalandi culture cages, mainly composed of the diatom
N. phyllepta and bacteria of the
Rhodobacteraceae family, demonstrated the ability to control the proliferation of unwanted bacteria, especially when in combination with probiotic microorganisms, and to be a valuable nutritional source for the fish, being rich in carbohydrates. Biofilms can also be used as indicators of water quality because their composition and structure are susceptible to temperature increases and ocean acidification [
65].
7. Conclusions
The interactions between marine diatoms and bacteria can have a substantial influence on ecosystem functioning, as they affect primary production, phytoplankton aggregation, and carbon and nutrient fluxes. Although the relevance of this interplay is gaining increasing attention from the scientific community, more studies are needed for a deeper understanding of the mechanisms involved. Due to the complexity of the interactions occurring, new approaches and methods are being developed to reveal the interactome network and identify compounds responsible for chemical communication among the organisms. The increasing level of knowledge in this field will have a pivotal role in establishing new approaches to answer questions related to environmental changes, but also in finding new solutions for a sustainable exploitation of diatom-microbiome consortia for biotechnological applications, from bioremediation strategies to the development of valuable products for human wellbeing.