1. Introduction
Retinoic acid (RA) is a metabolic product of Vitamin A, not inherently produced by the mammalian body and thus ingested from external sources in the form of carotenoids and retinyl esters from plants and animals, respectively. Vitamin A was first discovered in 1913 [
1], and numerous studies have demonstrated the necessity of this factor in early development, particularly in the development of the brain and vision [
2,
3]. The identification of the chemical structure can be attributed to Paul Karrer (1937, Nobel Prize in Chemistry) while the discovery of All-trans-Retinal as a crucial component in rhodopsin and thus subsequently necessary for vision is credited to George Wald (1967, Nobel Prize in Physiology and Medicine) [
4]. Vitamin A, taken up by the body, is stored in the liver primarily but can also be found in extrahepatic sites such as the lungs, bone marrow, and kidneys [
5].
2. Retinoic Acid Uptake and Metabolism
RBP4 (retinol-binding protein 4) is a protein found in the plasma capable of binding retinol and transporting it through the bloodstream to target cells containing the signaling receptor and transporter of retinol (STRA6) receptor [
6]. Once retinol enters the cytoplasm, it binds RBP1 (retinol-binding protein 1), to be stored before its transformation into All-trans retinoic acid (AtRA).
Retinol is converted to retinaldehyde via alcohol dehydrogenases (ADH) and retinol dehydrogenases (RDH), and then to retinoic acid via retinaldehyde dehydrogenases (RALDH) [
7]. The ADH family of enzymes can be ubiquitously expressed in embryonic and adult tissue (ADH5) or restricted to certain tissues (ADH1, ADH7) [
8]. Knockout studies suggest that ADH enzymes are most likely involved in curbing retinol toxicity than participating in retinoic acid synthesis [
9,
10]. RDH enzymes, on the other hand, have a more pronounced effect in optical structures [
11]. The RALDH family of enzymes consists of three enzymes—RALDH1, RALDH2, and RALDH3. Their distribution is site-specific and the availability of these enzymes serves as the rate-limiting step in retinoic acid synthesis. RALDH2, the earliest one to be expressed, first found in the primitive streak and mesodermal cells, is then localized to the somitic and lateral mesoderm, posterior heart tube, and rostral forebrain [
12]. It is responsible for retinoic acid availability up to ~E8.5, after which RALDH1 and RALDH3 contribute to retinoic acid synthesis in the eyes and the olfactory system.
Retinoic acid resembles fatty acids due to the presence of a beta-ionone ring, a polyunsaturated side chain, and a polar end group [
13], allowing for it to enter the cell easily through the plasma membrane. Now, in either an autocrine or paracrine fashion, retinoic acid diffuses into the neighboring cells, binds cellular retinoic acid-binding protein II (CRABPII), and moves into the nucleus. This is the point at which retinoic acid can now bind the retinoic acid receptors already bound to the DNA. After this action, retinoic acid moves out of the nucleus and is degraded by the CYP26 class of the cytochrome P450 (CYP450) superfamily of enzymes.
3. Retinoic Acid Receptors
Retinoic acid binds Retinoic Acid Receptor/Retinoid X Receptor (RAR/RXR) heterodimers, which are usually bound to retinoic acid response elements (RAREs) or retinoic X response elements (RXREs) in the promoter or enhancer regions of target genes [
14]. RAREs usually consist of two direct repeats of either the hexameric sequence (A/G)G(G/T)TCA or the more relaxed motif (A/G)G(G/T)(G/T)(G/T)(G/C)A, separated by 1, 2, or 5 bp (DR1, DR2, and DR5 RAREs, respectively) [
15]. This constitutive binding allows for the recruitment of corepressor complexes maintaining target gene repression.
The RAR and RXR receptors each exist as isotypes, RARα/β/γ and RXRα/β/γ, encoded by different genes, and each of them has its own isoforms, issued from alternative splicing and/or alternative promoter usage. While RXR receptors are only able to bind 9-cis retinoic acid (9-cis RA), RAR receptors can bind with high affinity AtRA and 9-cis RA. Each isotype receptor can be activated with specific ligands, that can be synthetic or naturally occurring. Natural agonists for RXR receptors are Docosahexaenoic acid, Lithocholic acid, and Methoprene acid [
16,
17]. There is an extensive list of pan-RXR synthetic agonists, including Bexarotene, LG100268, AGN194204, LG 101506, HX 630, and SR 11237. Phytanic acid, Honokiol, Peretinoin, AM-6-36, and CD3254 are RXRα specific agonists, while Danthron, derived from Chinese rhubarb, inversely is an RXRα specific antagonist [
18]. Conversely, for RAR receptors, several synthetic ligands have been designed; for instance, Am 580, Am 80, BMS614, BMS753, AGN 193835, and AGN 193836 are RARα-specific. BMS641 and 2-thienyl-substituted dihydronaphthalene retinoids are modified ligands that are RARβ-specific, while BMS961 and α-hydroxyacetamide-linked retinoids have been described as potent for RARγ activity, and TTNPB as a pan-RAR agonist [
17,
19].
Interestingly, the capacity of each of the synthetic RAR agonists to regulate a specific transcriptional response has been described early on by Pierre Chambon’s team [
20,
21,
22], and more recently, we have disentangled such a specific response at the level of the various controlled gene programs [
23,
24,
25].
4. The Role and Distribution of Retinoic Acid in Embryogenesis
The distribution of the RARs in embryos differs from region to region. Numerous studies looked into the expression patterns of the RAR and RXR receptors, especially in early mouse embryogenesis [
26,
27,
28], while a fair few also made comparisons with zebrafish [
29,
30,
31] and xenopus models [
32,
33,
34,
35]. In these studies, the presence of these receptors was systematically addressed by taking into consideration the developmental stages described from a chronological point of view (e.g., pregastrulation, gastrulation, neurula, and organogenesis [
36]).
Notably, while pregastrulation in mice and rat models does not reveal detectable levels of RAR/RXR gene expression, at gastrulation (E7.5), RARα and RARγ expression is ubiquitous and diffuse, and RARβ expression was found mainly in the lateral regions of the embryo [
28]. Similarly, RXR expression during pregastrulation is also not defined, with RAR and RXR expression often overlapping and omnipresent throughout the embryo.
In addition to the presence of RAR/RXR receptors, the driving differentiation force for site-specific cell specialization leading to development structures depends on the bioavailability of RA. Retinoic acid is first detected at the primitive streak phase, i.e., E7.5 of embryonic development in mice, confirmed by the expression of class IV ADH mRNA seen initially in the posterior region along the primitive streak [
8,
12]. It is then found to be localized close to the trunk, hindbrain, and optic vesicle by E8.5–E9.5 [
8,
37].
Postgastrulation is characterized by the regional patterning of the neurectoderm, the formation of the somites, the migration of cranial neural crest cells, and more. During this span of time, RAR expression becomes more distinctive. As summarized in
Figure 1A, by E8.5 to E13.5, there is an increased presence of RARα in the neuroectodermal region and in the regions distal to the caudal neuropore (CNP), as well as certain regions of the hindbrain up to rhombomere 6–7 (red line shading,
Figure 1A). RARβ is highly expressed in early stage midbrain at E8.5, specific to the neural tube and the rostral regions of the mesodermal tissue, and then, later on, in the spinal cord [
38,
39,
40] (green line shading;
Figure 1A). RARγ transcripts, on the other hand, occupy the opposite end, being found mostly in the regressing primitive streak and in the tail region of the somite-axis [
28], and almost completely absent from mesodermal tissue, as illustrated in the schematic representation provided in
Figure 1A (blue line shading).
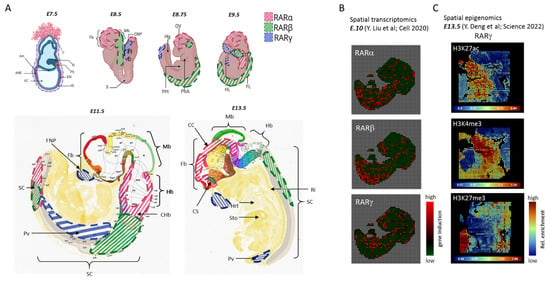
Figure 1. CNS-relevant RAR isotype transcript expression in mouse embryogenesis. (
A) Schematic representation of the expression of RARα (red line shading), RARβ (green line shading), and RARγ (blue line shading) transcripts at different stages of mouse embryogenesis from E7.5 to E13.5, particularly in relation to CNS development. At E7.5, very low to undetectable levels of RAR expression are observed. RARα is first seen at E8.5, lowly expressed in the neuroectoderm, near to the caudal neuropore (CNP). By E11.5, it is more present in the posterior region of the spinal cord (SC) and in the caudal hindbrain (CHb), before being expressed primarily in the corpus striatum (CS) and corpus callosum (CC) at E13.5. RARβ (green line shading) is present mostly in the neural tube between E8.5 and E9.5, with a more localized presence at the caudal region of the spinal cord (SC) at E11.5 onwards, while RARγ (blue line shading) is first seen in the regressing primitive streak (PS; at E8.5), and it is undetectable later on in the brain and spinal cord (SC) neuroepithelium. Its expression is limited to the frontonasal process (FNP) and branchial arches (limb region; HL, FL). Top panel created on Biorender.com. Bottom panel (E11.5, E13.5) images taken from Allen Developing Mouse Brain Reference Atlases (
https://developingmouse.brain-map.org/static/atlas) and overlayed with red/green/blue line shading to indicate RARα/β/γ expression, respectively. References made from studies [
8,
28,
37,
38,
39,
40,
41]. AC: Amniotic cavity, Al: Allantois, Am: Amnion, ANE: Anterior neuroectoderm, C: Chorion, CC: Corpus callosum, CHb: Caudal hindbrain, CNP: Caudal neuropore, CS: Corpus striatum, EM: Embryonic mesoderm, Fb: Forebrain, FL: Forelimb, FNP: Frontonasal process, Hb: Hindbrain, Hf: Headfold, Hg: Hindgut, HL: Hindlimb, Hrt: Heart, Mb: Midbrain, OV: Optic vesicle, PhA: Pharyngeal arch, PS: Primitive streak, Pv: Prevertebrae, Ri: Ribs, S: Somites, Sto: Stomach, VE: Visceral endoderm. (
B) RAR isotype over-expression revealed in spatial transcriptomics assay assessed in a
E.10 mouse embryo. The original data were produced by Liu et al. [
42], and the visualization was performed within MULTILAYER [
43,
44]. (
C) Histone modifications enrichment assessed within the gene coding region for the RARγ isotype, revealed by a spatial epigenomics assay performed on a E13.5 mouse embryo. Notice the coincident enrichment of the active marks H3K27ac and H3K4me3 within the frontonasal process region, as well as the enrichment of the repressive mark H3K27me3 in the developing CNS. The original data were produced by Deng et al. [
45], and displayed in AtlasXplore [
46].
By E13.5, RARα is majorly expressed in the corpus callosum and corpus striatum. At no point is RARγ (blue line shading) detected in the developing central nervous system (CNS). Its expression seems to be strongly repressed in this tissue layer, only appearing at the regions of frontonasal process (FNP), limb bud regions (HL, FL), and prevertebrae (Pv) from E9.5 onwards [
40]. This summarized view concerning the distribution of RARs during embryo development is further supported with recent high-throughput data obtained by spatial technologies, like spatial transcriptomics or spatial epigenomics. These methods allow researchers to capture localized gene expression or histone modification enrichment signatures within a tissue section. Specifically, the team of Dr. Rong Fan at Yale University developed a microfluidics system allowing to transport molecular biology reagents in a grid-based manner on top of a tissue section, thus allowing distinct molecular barcodes to be ligated to either cDNA or chromatin cleaved regions associated with specific histone modification marks [
42,
45]. This methodology has been evaluated on mouse embryos, which allowed us to query for molecular signatures associated with RAR isotypes. Indeed, the preferential RARβ expression along the spinal cord is fully recapitulated in spatial transcriptomics assays (
Figure 1B: E.10; Y. Liu et al. [
42]), and the described repression of RARγ in the developing CNS is fully confirmed by spatial epigenomics profiling targeting the histone modification H3K27me3, the most well-characterized gene silencing marker [
47]. Similarly, the RARγ preferential expression at the frontonasal process (FNP), displayed in
Figure 1A, correlates with the presence of the active marks H3K4me3 and H3K27ac, which concomitantly are known to create an open chromatin environment permissible to transcription [
47] (
Figure 1C: E13.5; Deng et al. [
45]).
Retinoic acid functionality also works in synchrony with complementary signals such as fibroblast growth factor (FGF-8), sonic hedgehog (SHH), and bone morphogenetic proteins (BMPs). Retinoic acid plays a role primarily in the growth and differentiation of the posterior structures, while in the anterior part of the embryo, its activity is regulated by the RA-degrading CYP450 enzymes CYP26A1 and CYP26C1, promoting the anterior-posterior patterning [
48,
49,
50]. Retinoic acid patterns the anteroposterior axis by being synthesized in the posterior mesoderm. The presence of CYP26C1 in the anterior mesoderm prevents the influence of retinoic acid in these regions, leading to the emergence of the hindbrain from the neural plate. In the dorsoventral axis of the developing neural tube, retinoic acid is produced at the somite sites, along with SHH and BMP expressed ventrally and dorsally, respectively. FGF-8 expression is detectable at the posterior end of the extending neural tube, and these factor gradients predict the ultimate fate of specialized neurons that emerge such as interneurons, sensory neurons, and motor neurons [
51,
52,
53].
This entry is adapted from the peer-reviewed paper 10.3390/life13122279