You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
In computer vision and image analysis, Convolutional Neural Networks (CNNs) and other deep-learning models are at the forefront of research and development. These advanced models have proven to be highly effective in tasks related to computer vision. One technique that has gained prominence in recent years is the construction of ensembles using deep CNNs. These ensembles typically involve combining multiple pretrained CNNs to create a more powerful and robust network.
- convolutional neural networks
- ensembles
- fusion
1. Introduction
Artificial neural networks (ANNs), which were initially developed in the 1950s, have had a checkered history, at times appreciated for their unique computational capabilities and at other times disparaged for being no better than statistical methods. Opinions shifted about a decade ago with deep neural networks, whose performance swiftly overshadowed that of other learners across various scientific (e.g., [1][2]), medical (e.g., [3][4]), and engineering domains (e.g., [5][6]). The prowess of deep learners is especially exemplified by the remarkable achievements of Convolutional Neural Networks (CNNs), one of the most renowned and robust deep-learning architectures.
CNNs have consistently outperformed other classifiers in numerous applications, particularly in image-recognition competitions where they frequently emerge as winners [7]. Not only do CNNs surpass traditional classifiers, but they also often outperform the recognition abilities of human beings. In the medical realm, CNNs have demonstrated superior performance compared to human experts in tasks such as skin cancer detection [8][9], identification of skin lesions on the face and scalp, and diagnosis of esophageal cancer (e.g., [10]). These remarkable achievements have naturally triggered a substantial increase in research focused on utilizing CNNs and other deep-learning techniques in medical imaging.
For instance, deep-learning models have emerged as the state-of-the-art for diagnosing conditions like diabetic retinopathy [11], Alzheimer’s disease [12], skin detection [13], gastrointestinal ulcers, and various types of cancer, as demonstrated in recent reviews and studies (see, for instance, [14][15]). Enhancing performance within the medical field carries the greater real impact of this technology compared to other applications.
CNNs, however, have limitations. It is widely recognized that they require many samples to avoid overfitting [16]. Acquiring image collections numbering in the hundreds of thousands for proper CNN training is an enormous enterprise [17]. In certain medical domains, it is prohibitively labor-intensive and costly [18]. Several well-established techniques have been developed to address the issue of overfitting with limited data, the two most common being transfer learning using pretrained CNNs and data augmentation [19][20]. The literature is abundant with studies investigating both methods, and it has been observed that combining the two yields better results (e.g., [21]).
In addition to transfer learning and data augmentation, another powerful technique for enhancing the performance of deep learners generally, as well as on small sample sizes, is to construct ensembles of pretrained CNNs [22]. Ensemble learning is a powerful technique in machine learning that aims to enhance predictive performance by combining the outputs of multiple classifiers [23]. The fundamental idea behind ensemble learning is to introduce diversity among the individual classifiers so that they collectively provide more accurate and robust predictions. This diversification can be achieved through various means, each contributing to the ensemble’s overall effectiveness [24].
One common approach to creating diversity among classifiers is to train each classifier on different subsets or variations of the available data [25]. This approach, known as data sampling or bootstrapping, allows each classifier to focus on different aspects or nuances within the dataset, which can lead to improved generalization and robustness. Another technique for introducing diversity is to use different types of CNN architectures within the ensemble [26]. By combining CNNs with distinct architectural features, such as varying kernel sizes, filter depths, or connectivity patterns, the ensemble can capture different aspects of the underlying data distribution, enhancing its overall predictive power [27][28]. In addition to varying architectural aspects, ensemble diversity can also be achieved by modifying network depth. Some classifiers within the ensemble may have shallower network architectures, while others may be deeper. This diversity in network depth can help the ensemble address different levels of complexity within the data, improving its adaptability to varying patterns and structures.
Furthermore, the ensemble can introduce diversity by using different activation functions within the neural networks. Activation functions play a crucial role in determining how information flows through the neural network layers. By employing a variety of activation functions, the ensemble can capture different types of non-linear relationships in the data, enhancing its ability to model complex patterns. Figure 1 depicts an example of a neural network in which each layer adopts an activation function that could be chosen at random among a set of available ones. The chosen activation function is then used by all the neurons in that layer.
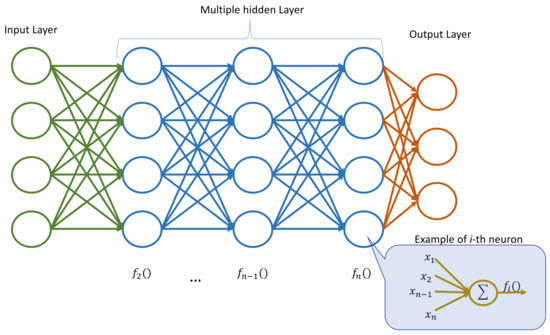
Figure 1. Example of neural network with multiple hidden layers. Each layer adopts a (possibly) different activation function to be used by all the neurons in that layer.
2. Methods for Building Ensembles of Convolutional Neural Networks
The related work in this field explores various strategies for creating ensembles of CNNs, with a focus on achieving high performance and maximizing the independence of predictions. Already addressed in the introduction are approaches based on training networks with different architectures and activation functions and using diverse training sets and data-augmentation approaches for the same network architecture. In addition, ensembles can be generated by combining multiple pretrained CNNs, employing various training algorithms, and applying distinct rules for combining networks.
The most intuitive approach to forming an ensemble involves training different models and then combining their outputs. Identifying the optimal classifier for a complex task can be a challenging endeavor [23]. Various classifiers may excel in leveraging the distinctive characteristics of specific areas within the given domain, potentially resulting in higher accuracy exclusively within those particular regions [29][30].
Most researchers taking this intuitive approach primarily fine-tune or train well-known architectures from scratch, average the results, and then demonstrate through experiments that the ensemble outperforms individual stand-alone networks. For instance, in [31], Kassani et al. employed an ensemble of VGG19 [32], MobileNet [33], and DenseNet [34] to classify histopathological biopsies, showing that the ensemble consistently achieved better performance than each individual network across four different datasets. Similarly, Qummar et al. [11] proposed an ensemble comprising ResNet50 [35], Inception v3 [36], Xception [37], DenseNet121, and DenseNet169 [34] to detect diabetic retinopathy.
In their study, Liu et al. [38] constructed an ensemble comprising three distinct CNNs proposed in their paper and averaged their results. Their ensemble achieved higher accuracy than the best individual model on the FER2013 dataset [39]. Similarly, Kumar et al. [40] introduced an ensemble of pretrained AlexNet and GoogleNet [41] models from ImageNet, which were then fine-tuned on the ImageCLEF 2016 collection dataset [42]. They utilized the features extracted from the last fully connected layers of these networks to train an SVM, an approach that outperformed CNN baselines and remained competitive with state-of-the-art methods at that time. Pandey et al. [43] proposed FoodNet, an ensemble composed of finetuned AlexNet, GoogleNet, and ResNet50 models designed for food image recognition. The output features from these models were concatenated and passed through a fully connected layer and softmax classifier.
Utilizing diverse training sets to train a classifier proves to be an effective approach to generating independent classifiers [23]. This can be achieved through various methods, with one classic technique being bagging [44][45][46]. Bagging involves creating m training sets of size n from a larger training set by randomly selecting samples with uniform probability and with replacement. Subsequently, the same model is trained on each of these training sets. Figure 2 depicts the bagging process. Examples of this approach to building ensembles include the work of Kim and Lim [47], who proposed a bagging-based approach to train three distinct CNNs for vehicle-type classification. Similarly, Dong et al. [48] applied bagging and CNNs to improve short-term load forecasting in a smart grid, resulting in a significant reduction of the mean absolute percentage error (MAPE) from 33.47 to 28.51. As another example, Guo and Gould [49] employed eight different datasets to train eight distinct networks for object detection. These datasets were formed by combining existing datasets in various ways. Remarkably, this straightforward approach led to substantial performance gains compared to individual models and brought the ensembles’s performance close to the state-of-the-art on competitive datasets like COCO 2012.
The training algorithms employed in CNNs follow stochastic trajectories and operate on stochastic data batches. Consequently, training the same network multiple times may lead to different models at the end of the process. Scholars can enhance the diversity among the final models in many ways: for instance, by employing different initialization of the initial model, or by adopting different optimization algorithms or loss functions during the training phase. Figure 3 depicts an example of an ensemble which adopt different optimization algorithms to introduce diversity. For instance, the authors in [50] constructed an ensemble for facial expression recognition using soft-label perturbation, where different losses were propagated for different samples. Similarly, Antipov et al. [51] utilized different network initializations to train multiple networks for gender predictions from face images.
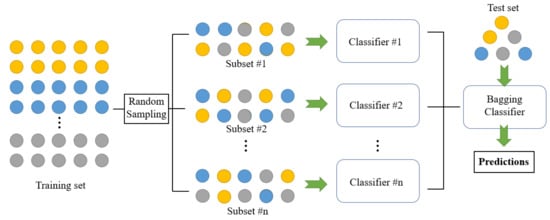
Figure 2. Structure of bagging classifier [52].
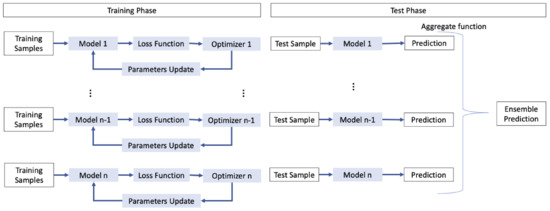
Figure 3. Example of ensemble that can use different optimization algorithms to introduce diversity.
Another approach to building ensembles is to adopt the same architecture but vary the activation functions. This can be done in a set of CNNs or within different layers of a single CNN [53]. One way to implement the latter approach is to select a random activation function from a pool for each layer in the original network [53].
Finally, ensembles can vary in the selection of rules for merging results. A straightforward approach is majority voting, where the predominant output selected by the majority of the networks is taken [54][55][56][57]. Another common technique frequently cited in the literature is to average the softmax outputs of the networks [58][59][60].
This entry is adapted from the peer-reviewed paper 10.3390/electronics12214428
References
- Wei, W.; Khan, A.; Huerta, E.; Huang, X.; Tian, M. Deep learning ensemble for real-time gravitational wave detection of spinning binary black hole mergers. Phys. Lett. B 2021, 812, 136029.
- Nanni, L.; Brahnam, S.; Lumini, A.; Loreggia, A. Coupling RetinaFace and Depth Information to Filter False Positives. Appl. Sci. 2023, 13, 2987.
- Shehab, M.; Abualigah, L.; Shambour, Q.; Abu-Hashem, M.A.; Shambour, M.K.Y.; Alsalibi, A.I.; Gandomi, A.H. Machine learning in medical applications: A review of state-of-the-art methods. Comput. Biol. Med. 2022, 145, 105458.
- Dutta, P.; Sathi, K.A.; Hossain, M.A.; Dewan, M.A.A. Conv-ViT: A Convolution and Vision Transformer-Based Hybrid Feature Extraction Method for Retinal Disease Detection. J. Imaging 2023, 9, 140.
- Wu, Z.; Tang, Y.; Hong, B.; Liang, B.; Liu, Y. Enhanced Precision in Dam Crack Width Measurement: Leveraging Advanced Lightweight Network Identification for Pixel-Level Accuracy. Int. J. Intell. Syst. 2023, 2023, 9940881.
- Deng, G.; Huang, T.; Lin, B.; Liu, H.; Yang, R.; Jing, W. Automatic meter reading from UAV inspection photos in the substation by combining YOLOv5s and DeepLabv3+. Sensors 2022, 22, 7090.
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25.
- Haggenmüller, S.; Maron, R.C.; Hekler, A.; Utikal, J.S.; Barata, C.; Barnhill, R.L.; Beltraminelli, H.; Berking, C.; Betz-Stablein, B.; Blum, A.; et al. Skin cancer classification via convolutional neural networks: Systematic review of studies involving human experts. Eur. J. Cancer 2021, 156, 202–216.
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118.
- Horie, Y.; Yoshio, T.; Aoyama, K.; Yoshimizu, S.; Horiuchi, Y.; Ishiyama, A.; Hirasawa, T.; Tsuchida, T.; Ozawa, T.; Ishihara, S.; et al. Diagnostic outcomes of esophageal cancer by artificial intelligence using convolutional neural networks. Gastrointest. Endosc. 2019, 89, 25–32.
- Qummar, S.; Khan, F.G.; Shah, S.; Khan, A.; Shamshirband, S.; Rehman, Z.U.; Khan, I.A.; Jadoon, W. A deep learning ensemble approach for diabetic retinopathy detection. IEEE Access 2019, 7, 150530–150539.
- Pan, D.; Zeng, A.; Jia, L.; Huang, Y.; Frizzell, T.; Song, X. Early detection of Alzheimer’s disease using magnetic resonance imaging: A novel approach combining convolutional neural networks and ensemble learning. Front. Neurosci. 2020, 14, 259.
- Nanni, L.; Loreggia, A.; Lumini, A.; Dorizza, A. A Standardized Approach for Skin Detection: Analysis of the Literature and Case Studies. J. Imaging 2023, 9, 35.
- Nagaraj, P.; Subhashini, S. A Review on Detection of Lung Cancer Using Ensemble of Classifiers with CNN. In Proceedings of the 2023 2nd International Conference on Edge Computing and Applications (ICECAA), Namakkal, India, 19–21 July 2023; pp. 815–820.
- Shah, A.; Shah, M.; Pandya, A.; Sushra, R.; Sushra, R.; Mehta, M.; Patel, K.; Patel, K. A Comprehensive Study on Skin Cancer Detection using Artificial Neural Network (ANN) and Convolutional Neural Network (CNN). Clin. eHealth 2023, 6, 76–84.
- Thanapol, P.; Lavangnananda, K.; Bouvry, P.; Pinel, F.; Leprévost, F. Reducing overfitting and improving generalization in training convolutional neural network (CNN) under limited sample sizes in image recognition. In Proceedings of the 2020-5th International Conference on Information Technology (InCIT), Chonburi, Thailand, 21–22 October 2020; pp. 300–305.
- Campagner, A.; Ciucci, D.; Svensson, C.M.; Figge, M.T.; Cabitza, F. Ground truthing from multi-rater labeling with three-way decision and possibility theory. Inf. Sci. 2021, 545, 771–790.
- Panch, T.; Mattie, H.; Celi, L.A. The “inconvenient truth” about AI in healthcare. NPJ Digit. Med. 2019, 2, 77.
- Bravin, R.; Nanni, L.; Loreggia, A.; Brahnam, S.; Paci, M. Varied Image Data Augmentation Methods for Building Ensemble. IEEE Access 2023, 11, 8810–8823.
- Claro, M.L.; de MS Veras, R.; Santana, A.M.; Vogado, L.H.S.; Junior, G.B.; de Medeiros, F.N.; Tavares, J.M.R. Assessing the Impact of Data Augmentation and a Combination of CNNs on Leukemia Classification. Inf. Sci. 2022, 609, 1010–1029.
- Nanni, L.; Fantozzi, C.; Loreggia, A.; Lumini, A. Ensembles of Convolutional Neural Networks and Transformers for Polyp Segmentation. Sensors 2023, 23, 4688.
- Nanni, L.; Lumini, A.; Loreggia, A.; Brahnam, S.; Cuza, D. Deep ensembles and data augmentation for semantic segmentation. In Diagnostic Biomedical Signal and Image Processing Applications with Deep Learning Methods; Elsevier: Amsterdam, The Netherlands, 2023; pp. 215–234.
- Cornelio, C.; Donini, M.; Loreggia, A.; Pini, M.S.; Rossi, F. Voting with random classifiers (VORACE): Theoretical and experimental analysis. Auton. Agent 2021, 35, 2.
- Yao, X.; Liu, Y. Making use of population information in evolutionary artificial neural networks. IEEE Trans. Syst. Man Cybern. Part B (Cybern.) 1998, 28, 417–425.
- Opitz, D.; Shavlik, J. Generating accurate and diverse members of a neural-network ensemble. Adv. Neural Inf. Process. Syst. 1995, 8.
- Liu, Y.; Yao, X.; Higuchi, T. Evolutionary ensembles with negative correlation learning. IEEE Trans. Evol. Comput. 2000, 4, 380–387.
- Rosen, B.E. Ensemble learning using decorrelated neural networks. Connect. Sci. 1996, 8, 373–384.
- Liu, Y.; Yao, X. Ensemble learning via negative correlation. Neural Netw. 1999, 12, 1399–1404.
- Matloob, F.; Ghazal, T.M.; Taleb, N.; Aftab, S.; Ahmad, M.; Khan, M.A.; Abbas, S.; Soomro, T.R. Software defect prediction using ensemble learning: A systematic literature review. IEEE Access 2021, 9, 98754–98771.
- Roshan, S.E.; Asadi, S. Improvement of Bagging performance for classification of imbalanced datasets using evolutionary multi-objective optimization. Eng. Appl. Artif. Intell. 2020, 87, 103319.
- Kassani, S.H.; Kassani, P.H.; Wesolowski, M.J.; Schneider, K.A.; Deters, R. Classification of histopathological biopsy images using ensemble of deep learning networks. arXiv 2019, arXiv:1909.11870.
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556.
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition , Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520.
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708.
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778.
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 2818–2826.
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258.
- Liu, K.; Zhang, M.; Pan, Z. Facial expression recognition with CNN ensemble. In Proceedings of the 2016 International Conference on Cyberworlds (CW), Chongqing, China, 28–30 September 2016; pp. 163–166.
- Goodfellow, I.J.; Erhan, D.; Carrier, P.L.; Courville, A.; Mirza, M.; Hamner, B.; Cukierski, W.; Tang, Y.; Thaler, D.; Lee, D.H.; et al. Challenges in representation learning: A report on three machine learning contests. In Proceedings of the Neural Information Processing: 20th International Conference, ICONIP 2013, Daegu, Republic of Korea, 3–4 November 2013; Proceedings, Part III 20. Springer: Cham, Switzerland, 2013; pp. 117–124.
- Kumar, A.; Kim, J.; Lyndon, D.; Fulham, M.; Feng, D. An ensemble of fine-tuned convolutional neural networks for medical image classification. IEEE J. Biomed. Health Inform. 2016, 21, 31–40.
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9.
- Gilbert, A.; Piras, L.; Wang, J.; Yan, F.; Ramisa, A.; Dellandrea, E.; Gaizauskas, R.J.; Villegas, M.; Mikolajczyk, K. Overview of the ImageCLEF 2016 Scalable Concept Image Annotation Task. In Proceedings of the CLEF (Working Notes), Évora, Portugal, 5–8 September 2016; pp. 254–278.
- Pandey, P.; Deepthi, A.; Mandal, B.; Puhan, N.B. FoodNet: Recognizing foods using ensemble of deep networks. IEEE Signal Process. Lett. 2017, 24, 1758–1762.
- Breiman, L. Bagging predictors. Mach. Learn. 1996, 24, 123–140.
- Wolpert, D.H.; Macready, W.G. An efficient method to estimate bagging’s generalization error. Mach. Learn. 1999, 35, 41–55.
- Bauer, E.; Kohavi, R. An empirical comparison of voting classification algorithms: Bagging, boosting, and variants. Mach. Learn. 1999, 36, 105–139.
- Kim, P.K.; Lim, K.T. Vehicle type classification using bagging and convolutional neural network on multi view surveillance image. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Honolulu, HI, USA, 21–26 July 2017; pp. 41–46.
- Dong, X.; Qian, L.; Huang, L. A CNN-based bagging learning approach to short-term load forecasting in smart grid. In Proceedings of the 2017 IEEE SmartWorld, Ubiquitous Intelligence & Computing, Advanced & Trusted Computed, Scalable Computing & Communications, Cloud & Big Data Computing, Internet of People and Smart City Innovation (SmartWorld/SCALCOM/UIC/ATC/CBDCom/IOP/SCI), San Francisco, CA, USA, 4–8 August 2017; pp. 1–6.
- Guo, J.; Gould, S. Deep CNN ensemble with data augmentation for object detection. arXiv 2015, arXiv:1506.07224.
- Gan, Y.; Chen, J.; Xu, L. Facial expression recognition boosted by soft label with a diverse ensemble. Pattern Recognit. Lett. 2019, 125, 105–112.
- Antipov, G.; Berrani, S.A.; Dugelay, J.L. Minimalistic CNN-based ensemble model for gender prediction from face images. Pattern Recognit. Lett. 2016, 70, 59–65.
- Zhang, H.; Zhou, T.; Xu, T.; Hu, H. Remote interference discrimination testbed employing AI ensemble algorithms for 6G TDD networks. Sensors 2023, 23, 2264.
- Nanni, L.; Lumini, A.; Ghidoni, S.; Maguolo, G. Stochastic selection of activation layers for convolutional neural networks. Sensors 2020, 20, 1626.
- Ju, C.; Bibaut, A.; van der Laan, M. The relative performance of ensemble methods with deep convolutional neural networks for image classification. J. Appl. Stat. 2018, 45, 2800–2818.
- Harangi, B. Skin lesion classification with ensembles of deep convolutional neural networks. J. Biomed. Inform. 2018, 86, 25–32.
- Lyksborg, M.; Puonti, O.; Agn, M.; Larsen, R. An ensemble of 2D convolutional neural networks for tumor segmentation. In Proceedings of the Image Analysis: 19th Scandinavian Conference, SCIA 2015, Copenhagen, Denmark, 15–17 June 2015; Proceedings 19. Springer: Cham, Switzerland, 2015; pp. 201–211.
- Minetto, R.; Segundo, M.P.; Sarkar, S. Hydra: An ensemble of convolutional neural networks for geospatial land classification. IEEE Trans. Geosci. Remote Sens. 2019, 57, 6530–6541.
- Dong, X.; Yu, Z.; Cao, W.; Shi, Y.; Ma, Q. A survey on ensemble learning. Front. Comput. Sci. 2020, 14, 241–258.
- Brown, G.; Wyatt, J.; Harris, R.; Yao, X. Diversity creation methods: A survey and categorisation. Inf. Fusion 2005, 6, 5–20.
- Sagi, O.; Rokach, L. Ensemble learning: A survey. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2018, 8, e1249.
This entry is offline, you can click here to edit this entry!
