1. Photoperiodic Pathway
The photoperiodic pathway, characterized by the differential stability and activity of
CONSTANS (
CO), plays a pivotal role in modulating flowering time.
CO, a rhythmically expressed gene, exhibits responsiveness to both circadian rhythms and light stimuli under varying daylengths [
41]. This dynamic regulation influences the accumulation of FT, thereby contributing to the precise temporal control of flowering events. Noncoding RNAs play a crucial regulatory role in photoperiodic flowering by influencing circadian rhythms or upstream factors of CO. For instance, miR397b targets
CKB3, impacting the activity of CCA1, a central component of the circadian clock. Overexpressing miR397b extends the circadian clock period, consequently delaying flowering. Conversely, miR397b is negatively regulated by CCA1, establishing a negative feedback loop of miR397b-
CKB3-CCA1 [
42]. The lncRNA
FLORE, a natural antisense transcript of
CDF5, inhibited the expression of
CDF5. CDFs, under circadian transcriptional and post-translational control, are repressors of
CO and
FT; thus, overexpression of
FLORE promotes flowering by upregulation of
CO and
FT [
41,
43]. The reciprocal inhibition of
FLORE and
CDF5 is crucial for maintaining their oscillations [
44].
Besides functioning upstream of
CO, evidence also shows that noncoding RNAs function downstream of
CO to regulate flowering. Overexpression of
RIFLA, an lncRNA transcribed from the intron of
OsMADS56, results in earlier flowering than the wild type under long-day conditions [
45].
OsMADS56 is the ortholog of the
Arabidopsis SOC1 in rice, positioned downstream of
CO, facilitating the integration of signals conveyed by CO to promote flowering in plants. However, in contrast to facultative long-day plants, overexpression of
OsMADS56 in short-day crop plants like rice delays flowering under long-day conditions. Additionally, miR5200 in
Brachypodium distachyon is epigenetically regulated by daylength and short-day induction, leading to delayed flowering by targeting
FTL1 and
FTL2 [
46]. However, the diurnal or circadian expression patterns of these genes have not been elucidated. Understanding their expression patterns would significantly contribute to our comprehension of their roles within the photoperiodic pathway, allowing for better utilization and manipulation of their functions.
2. Autonomous Pathway and Vernalization Pathway
Plants require exposure to low temperatures to induce flowering, a phenomenon known as vernalization. Vernalization induces the inactivation of
FLC, thereby promoting flowering. The autonomous pathway comprises the complex of
FLOWERING LOCUS CA (
FCA),
FLOWERING LOCUS PA (
FPA),
FLOWERING LOCUS KH DOMAIN (
FLK), and
FLOWERING LOCUS Y (
FY), responsible for repressing the expression of
FLC by modifying 3′ end processing and antisense RNA polyadenylation at
FLC [
47]. If the complex of the autonomous pathway is absent, even vernalized plants cannot flower early. Hence,
FLC occupies a central position between the vernalization and autonomous pathways [
48]. LncRNAs such as
COOLAIR,
COLDAIR, and
COLDWRAP are induced by cold treatment, mimicking winter conditions, and repress
FLC expression.
COOLAIR, an antisense lncRNA originating from the 3′ end of
FLC, promotes flowering by repressing
FLC expression [
49,
50].
COLDAIR is a sense lncRNA from the intron of
FLC, and perturbating the association between
COLDAIR and
FLC delays flowering time in
Arabidopsis [
51]. Another lncRNA,
COLDWRAP, derived from the promoter of
FLC, also contributes to the de-repression of flowering in coordination with
COLDAIR [
52]. In addition, FPA, FCA, and FY can also regulate the alternative splicing of
COOLAIR, recruiting FLD for H3K4me2 demethylation on
FLC. These findings suggest that FLC is intricately regulated by noncoding RNAs, involving both the vernalization and autonomous pathways [
47,
53]. However, the interplay between noncoding RNAs such as
COOLAIR,
COLDAIR, and
COLDWRAP in coordinating
FLC modifications and regulating
FLC expression requires further in-depth investigation. Additionally, datasets have shown that the flowering repressor genes
FLC and
FLM in Arabidopsis produce circRNAs [
34]. These findings indicate that
FLC, positioned at the intersection of the vernalization and autonomous pathways, is subject to regulation by various noncoding RNAs.
Besides the
FLC locus, long noncoding RNAs generated from other loci have also been reported to regulate the vernalization pathway. A natural antisense transcript (NAT) from
MADS AFFECTING FLOWERING 4 (
MAF4) called
MAS is also cold-induced and activates
MAF4 expression to inhibit premature flowering [
54]. Although the function of MAF4 appears to oppose the long noncoding RNAs generated from the
FLC locus, this ensures that the plant flowers only after experiencing a sufficiently long period of low temperatures. Additionally, there are other long noncoding RNAs that have been identified through transcriptome data screening. LncRNA
TCONS_00035129 in
Brassica rapa is induced by vernalization, positively correlating with
BraZF-HD21 [
55]. LncRNA
AGL15X2 in
Beta vulgaris L. reaches peak expression after 16 weeks of vernalization, promoting flowering by repressing
BvFT1 [
56]. An intergenic lncRNA,
FLINC from Arabidopsis, has been identified to play a role in temperature-mediated flowering. Overexpression of
FLINC increases sensitivity to changes in ambient temperature [
57]. However, how these long noncoding RNAs respond to vernalization and are induced for expression and the mechanistic understanding of these long noncoding RNAs remain incomplete, necessitating further molecular biology and genetic research for elucidation.
3. Aging Pathway
Plant age is a critical factor in regulating the transition to flowering, and research has indicated a close correlation between age and miR156/miR172. MiR156 exhibits higher expression levels in juvenility, gradually decreasing with age, crucial for maintaining the juvenile phase. Subsequent studies have demonstrated that the temporal expression of miR156 is regulated by sugars synthesized during photosynthesis. MiR156 targets the
SQUAMOSA promoter binding protein-like (
SPL) family, known as flowering-promoting factors, where the gradual downregulation of miR156 leads to a progressive increase in SPL expression. Overexpression of miR156 extends the juvenile stage and delays flowering in several plant species, including Arabidopsis, rice,
Zea mays ssp.
mays (maize),
Lycopersicon esculentum (tomato),
Populus ×
canadensis ,
Lilium ×
formolongi, and
Gossypium hirsutum L. (cotton), underscoring the conserved function of this miRNA family [
58,
59,
60,
61,
62]. Notably, in
Physcomitrella patens, miR156 exhibits an opposing role by promoting the formation of leafy gametophores [
63]. The SPL family encodes a group of transcription factors targeted by miR156. In Arabidopsis, miR156 targets 11 of 17
SPL members, with SPL3/4/5/9/15 being key contributors to flowering time regulation [
64]. Gradually increased SPL9, 10, and 15 can bind to the promoter region of
MIR172B, enhancing its expression and consequently downregulating the expression of the flowering repressor genes
APETALA2 (
AP2) and
AP2-like. Thus, miR172 acts downstream of miR156 and SPL9 and its abundance increases with plant age, exhibiting an expression pattern opposite to miR156. Overexpressing miR172 accelerates flowering in plants, contrasting the effects of miR156 [
65]. Target analysis reveals that miR172 represses
AP2,
SMZ,
SNZ, and
TOE1/2/3 [
23,
66,
67]. Therefore, miR156-
SPLs-miR172 defines an age pathway in a wide range of plants.
It is interesting that some other noncoding RNAs are predicted to target miR156 and miR172, acting as decoys to inhibit their binding to their respective target genes. For example, circ-CCR2, circ-SEC5A, and circ-EF1B may be involved in aging pathways, as they sponge miR156 or miR172 in soybean [
38]. However, whether these circRNAs have an impact on aging and their actual role in regulating flowering time remains to be studied. Recently, heterologous expression of lncRNA
bra-miR156HG from
Brassica campestris (Chinese cabbage), which is assumed to be the precursor of miR156, leads to delayed flowering in Arabidopsis. However, overexpression of this lncRNA alters leaf morphology instead of changing flowering time in Chinese cabbage [
68]. This indicates that various types of noncoding RNAs collaborate to regulate flowering in a single pathway, a phenomenon that is relatively scarce in plants currently.
Furthermore, age can also regulate miRNA splicing in a post-transcriptional manner. The splicing isoforms of miR528 are regulated by age.
Pri-miR528 has two splicing variants,
MIR528-T1 and
MIR528-T2. In
MIR528-T1, the 3′ end is shortened by 103 nucleotides due to proximal polyadenylation, while in
MIR528-T2, it is shortened by 98 nucleotides due to intron splicing, although the mature sequence of miR528 is the same. Older plants tend to produce the
MIR528-T2 isoform, which weakens the ability to yield mature miR528, which promotes flowering by targeting
OsRFI2 [
69]. This age-regulated splicing of miR528 represents a fine-tuning mechanism, enhancing its ability to promote flowering just before the reproductive transition. Further investigation is required to elucidate which factor regulates the splicing of miR528 and whether it is correlated with miR156 or miR172.
3.4. Phytohormone-Related Pathways
GA was initially considered the primary plant hormone regulating flowering, and its pathway has been extensively studied. DELLA proteins function as negative regulators in the GA signaling pathway, inhibiting the floral integrator factor
SOC1 and thereby impeding flowering. GA promotes the degradation of DELLA proteins, relieving SOC1 inhibition and promoting flowering [
70]. MiR159 targets
MYB transcription factors responsive to GA [
71]. However, the inconsistencies in flowering phenotypes between miR159 and target
MYB mutants suggest the involvement of other factors in this regulatory module [
72].
Other plant hormone signaling components may indirectly regulate flowering by interacting with DELLA proteins. MiR390-
TAS3-tasiRNAs participate in auxin-mediated flowering by targeting
auxin response factor (
ARF)
3/4 [
73]. ARFs are negatively regulated by the auxin receptor Aux/IAA, which can indirectly modulate flowering by promoting GA20ox or inhibiting GA2ox to increase GA levels [
74]. Additionally, Aux/IAA negatively regulates DELLA proteins, demonstrating the collaborative role of auxin and GA in flowering [
75]. Similarly, OsmiR393 targets the auxin receptors
OsTIR1 and
OsAFB2, and overexpression of miR393 affects auxin signaling, thereby impacting flowering time [
76].
Brassinosteroids (BRs) are a group of polyhydroxylated steroidal hormones crucial in plant growth and development. Mutants lacking BRs exhibit delayed flowering, a phenotype restored by exogenous GA supplementation, demonstrating the synergistic action between BRs and GA [
77]. Additionally, BR signaling can function independently of GA. In
Arabidopsis, the BR signaling component BZR1 recruits the H3K27 demethylase ELF6 to the
FLC gene locus, inducing H3K27 demethylation and consequently activating
FLC expression and promoting flowering [
78]. In rice, although there is no homologous gene to
FLC [
79], a study has indicated that overexpression of OsmiR397 enhances the plant’s BR signal, with an earlier heading time [
12]. While the exact cause remains unclear, this suggests the potential existence of an alternative mechanism for BR-mediated flowering regulation in rice.
Additionally, some noncoding RNA-mediated pathways regulating flowering remain unclear or challenging to categorize into specific pathways. For instance, in rice, overexpression of miR168 can elevate miR164 levels, delaying flowering [
80]. MiR164 targets the transcription factors
CUC1 and
CUC2, which are crucial for the formation of floral organ boundaries [
81].
Ef-cd is an antisense lncRNA overlapping with
OsSOC1, and it shortens the maturity duration, thereby accelerating heading in rice [
82]. This example is challenging to categorize within the mentioned pathways since
SOC1 is a gene downstream in the process, where all pathways converge to trigger flowering. Recently, an intergenic lncRNA,
FLAIL, was identified, and its mutants displayed an early flowering phenotype, indicating that
FLAIL acts as a flowering repressor [
30]. Mechanistic studies indicate that
FLAIL reduces the expression of
LAC8 by influencing alternative splicing, consequently altering the flowering time. Interestingly,
LAC8 belongs to the laccase family along with the OsmiR397 target gene
OsLAC13, hinting at potential flowering control pathways associated with laccase-related mechanisms in plants [
12]. In citrus, overexpression of miR3954 promotes the production of phasiRNAs that target NAC genes, facilitating the regulation of flowering time [
43].
In
Arabidopsis, a circRNA derived from the intron of
At5g37720 significantly delays flowering time, with reduced expression of
FT [
58]. However, its mechanism of action remains unclear, potentially involving the circRNA binding to certain proteins and exerting trans-regulation over
FT expression. Recently, the loss of function of miR394a and miR394b in Arabidopsis resulted in early flowering with reduced branching and lower seed production [
41]. However, the phenotypic alteration in flowering time does not correlate with the known target gene
LCR. Therefore, the mechanism by which miR394 regulates flowering remains unknown. Further in-depth research in the future will help us comprehend these unknown regulatory mechanisms. It may establish connections with known pathways or create novel pathways to explain these phenomena.
In summary, noncoding RNAs intricately modulate plant flowering time through the complex web of pathways described above (Figure 1), although the mechanisms of some newly identified flowering-related noncoding RNAs are yet to be fully explored. The multifaceted involvement of noncoding RNAs underscores their pivotal role in the regulation of flowering time across various plant species (Table 1).
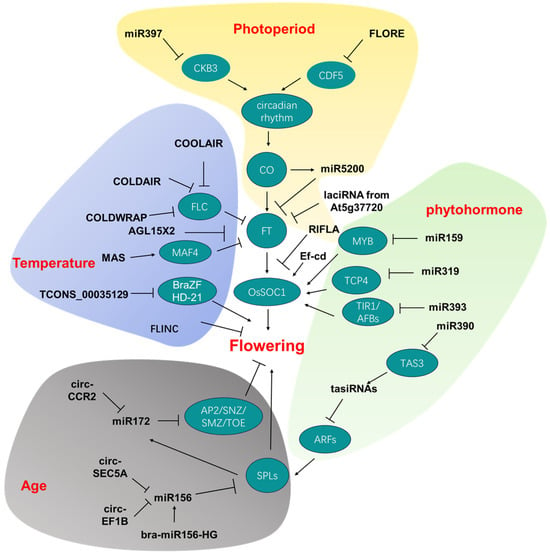
Figure 1. Noncoding RNAs involved in flowering time and their targets/downstream genes. Different colors of background represent different pathways, indicated by the word in red. CKB3: Casein Kinase Beta subunit 3; CDF5: CYCLING DOF FACTOR 5; CO: CONSTANS; FT: FLOWERING LOCUS T; COOLAIR: COLD INDUCED LONG ANTISENSE INTRAGENIC RNA; COLDAIR: COLD-ASSISTED INTRONIC NONCODING RNA; FLORE: CDF5 LONG NONCODING RNA; FLC: FLOWERING LOCUS C; MAF4: MADS AFFECTING FLOWERING 4; BraZF-HD21:Bra026812; MYB: v-myb avian myeloblastosis viral oncogene homolog transcription factor; TCP: TEOSINTE BRANCHED 1, CYCLOIDEA, POLIFERATING CELL FACTORS; TIR1:TRANSPORT INHIBITOR RESISTANT1; AFB:AUXIN SIGNALING F-BOX; TAS3: Trans-Acting Short Interference RNA3; ARF: auxin response factor; SPL: SQUAMOSA promoter binding protein-like; AP2: APETALA2 transcription factors; SNZ: SCHNARCHZAPFEN; SMZ: SCHLAFMUTZE;TOE: TARGET OF EAT.
This entry is adapted from the peer-reviewed paper 10.3390/genes14122114