CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 is a unique genome editing tool that can be easily used in a wide range of applications, including functional genomics, transcriptomics, epigenetics, biotechnology, plant engineering, livestock breeding, gene therapy, diagnostics, and so on.
- genome editing
- CRISPR/Cas9
- therapeutics
- diagnostics
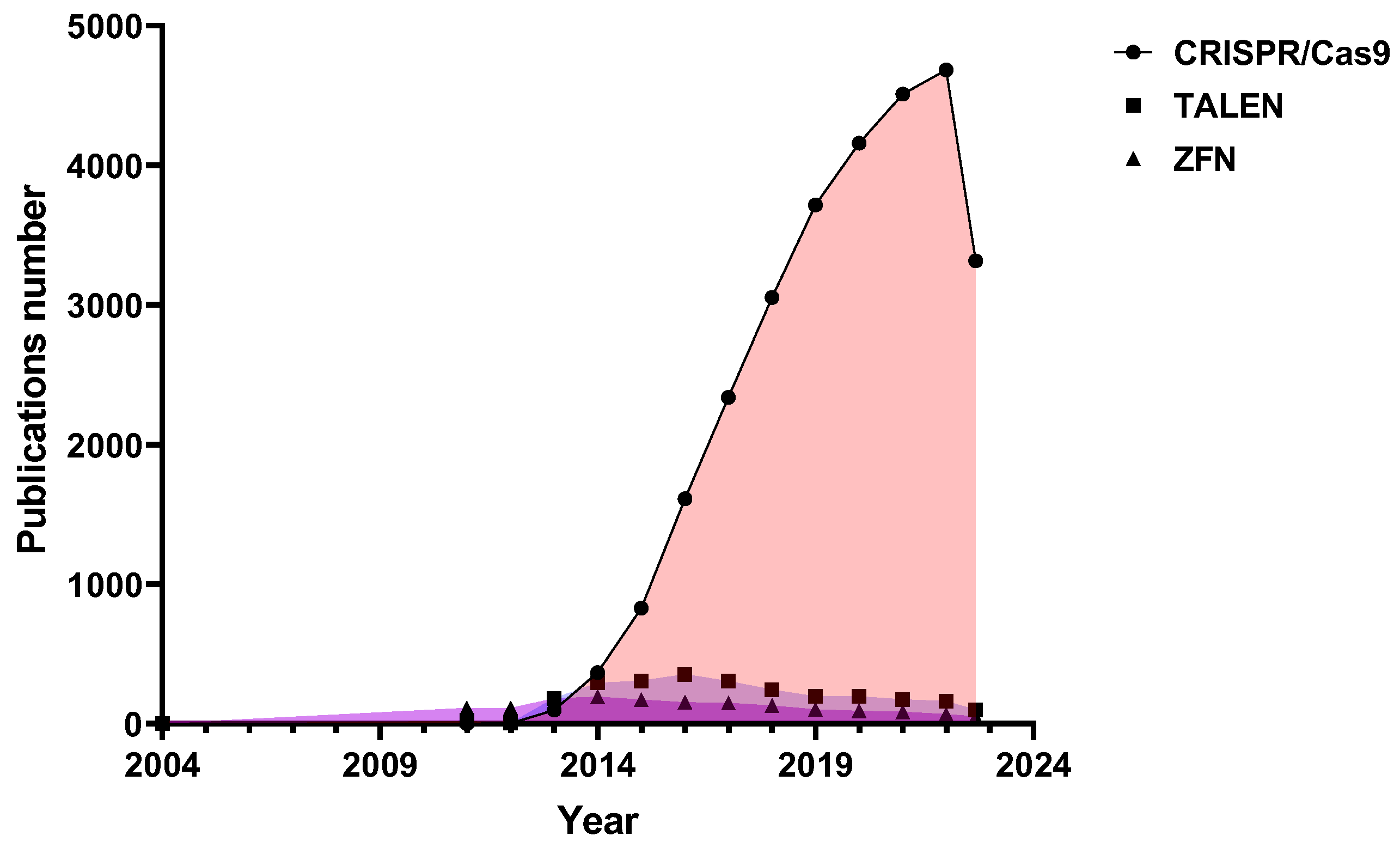
1. Introduction
2. CRISPR/Cas9 Delivery Methods
|
Delivery Approach |
Mode of Cas9 and Guide RNA Delivery |
||
|---|---|---|---|
|
DNA |
mRNA |
RNP |
|
|
Electroporation |
+ |
+ |
+ |
|
Viral vectors |
+ |
+ |
− |
|
Lipofection |
+ |
+ |
+ |
|
Lipid nanoparticles |
− |
+ |
+ |
|
Polymer nanoparticles |
− |
− |
+ |
|
Hydrogel nanoparticles |
− |
− |
+ |
|
Gold nanoparticles |
− |
− |
+ |
|
Graphene oxide |
− |
− |
+ |
|
Metal−organic frameworks |
− |
− |
+ |
|
Black phosphorus nanosheets |
− |
− |
+ |
|
Cell-penetrating peptides |
− |
− |
+ |
|
DNA nanostructures |
− |
− |
+ |
3. CRISPR/Cas9-Based Diagnostics
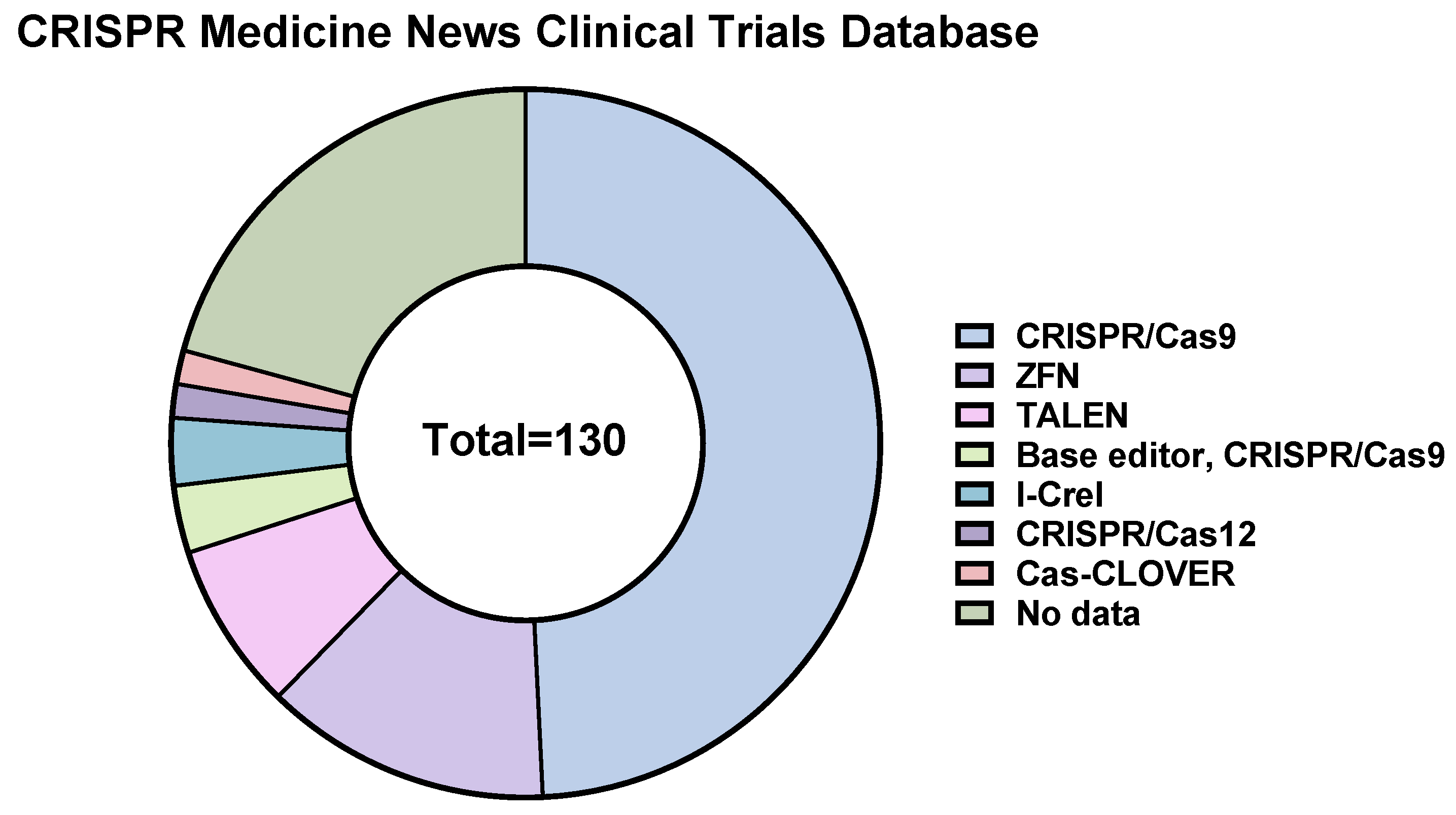
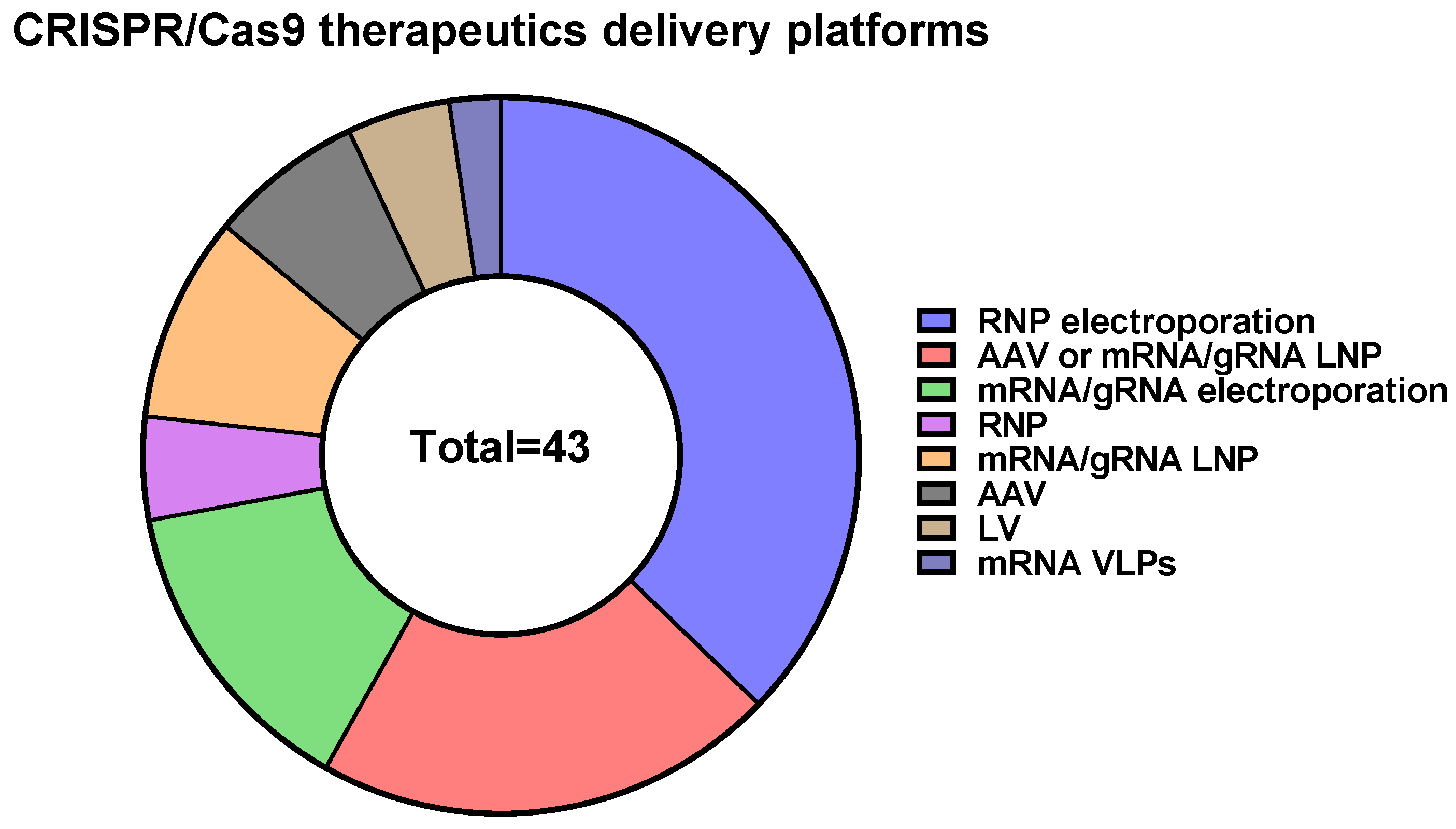
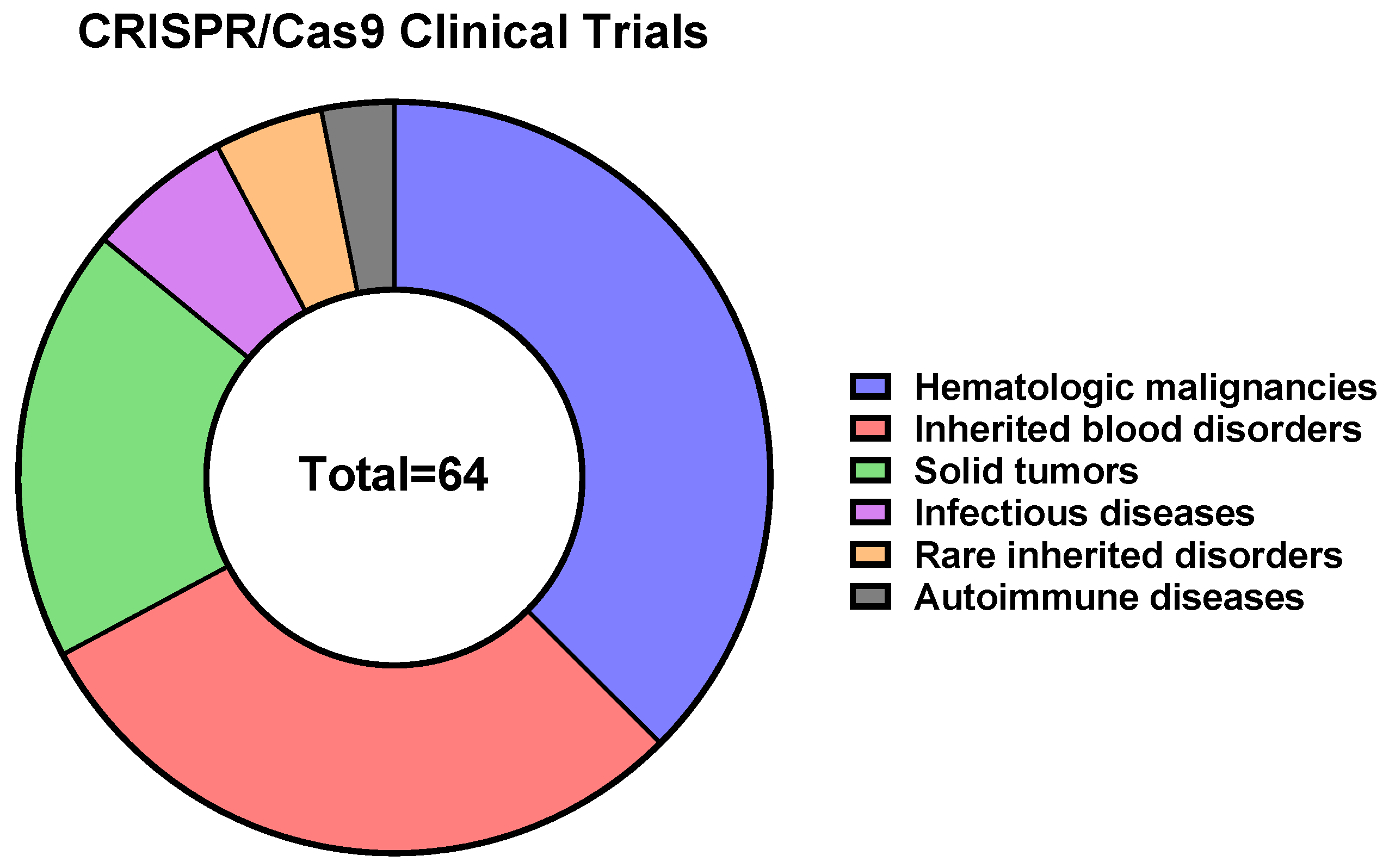
4. CRISPR/Cas9-Based Therapeutics
This entry is adapted from the peer-reviewed paper 10.3390/ijms242216077
References
- Gaj, T.; Sirk, S.J.; Shui, S.-L.; Liu, J. Genome-Editing Technologies: Principles and Applications. Cold Spring Harb. Perspect. Biol. 2016, 8, a023754.
- Li, H.; Yang, Y.; Hong, W.; Huang, M.; Wu, M.; Zhao, X. Applications of Genome Editing Technology in the Targeted Therapy of Human Diseases: Mechanisms, Advances and Prospects. Signal Transduct. Target. Ther. 2020, 5, 1.
- Matsumoto, D.; Nomura, W. The History of Genome Editing: Advances from the Interface of Chemistry & Biology. Chem. Commun. 2023, 59, 7676–7684.
- Bock, C.; Datlinger, P.; Chardon, F.; Coelho, M.A.; Dong, M.B.; Lawson, K.A.; Lu, T.; Maroc, L.; Norman, T.M.; Song, B.; et al. High-Content CRISPR Screening. Nat. Rev. Methods Primers 2022, 2, 8.
- Grav, L.M.; la Cour Karottki, K.J.; Lee, J.S.; Kildegaard, H.F. Application of CRISPR/Cas9 Genome Editing to Improve Recombinant Protein Production in CHO Cells. In Methods in Molecular Biology; Springer: New York, NY, USA, 2017; pp. 101–118. ISBN 9781493969715.
- Chan, K.F.; Shahreel, W.; Wan, C.; Teo, G.; Hayati, N.; Tay, S.J.; Tong, W.H.; Yang, Y.; Rudd, P.M.; Zhang, P.; et al. Inactivation of GDP-fucose Transporter Gene (Slc35c1) in CHO Cells by ZFNs, TALENs and CRISPR-Cas9 for Production of Fucose-free Antibodies. Biotechnol. J. 2016, 11, 399–414.
- Geurts, A.M.; Cost, G.J.; Freyvert, Y.; Zeitler, B.; Miller, J.C.; Choi, V.M.; Jenkins, S.S.; Wood, A.; Cui, X.; Meng, X.; et al. Knockout Rats via Embryo Microinjection of Zinc-Finger Nucleases. Science 2009, 325, 433.
- Lee, H.; Yoon, D.E.; Kim, K. Genome Editing Methods in Animal Models. Anim. Cells Syst. 2020, 24, 8–16.
- Lin, Y.; Li, J.; Li, C.; Tu, Z.; Li, S.; Li, X.-J.; Yan, S. Application of CRISPR/Cas9 System in Establishing Large Animal Models. Front. Cell Dev. Biol. 2022, 10, 919155.
- Gan, W.C.; Ling, A.P.K. CRISPR/Cas9 in Plant Biotechnology: Applications and Challenges. BioTechnologia 2022, 103, 81–93.
- Son, S.; Park, S.R. Challenges Facing CRISPR/Cas9-Based Genome Editing in Plants. Front. Plant Sci. 2022, 13, 902413.
- Singh, P.; Ali, S.A. Impact of CRISPR-Cas9-Based Genome Engineering in Farm Animals. Vet. Sci. 2021, 8, 122.
- Menchaca, A.; dos Santos-Neto, P.C.; Mulet, A.P.; Crispo, M. CRISPR in Livestock: From Editing to Printing. Theriogenology 2020, 150, 247–254.
- Wang, H.-X.; Li, M.; Lee, C.M.; Chakraborty, S.; Kim, H.-W.; Bao, G.; Leong, K.W. CRISPR/Cas9-Based Genome Editing for Disease Modeling and Therapy: Challenges and Opportunities for Nonviral Delivery. Chem. Rev. 2017, 117, 9874–9906.
- Kotterman, M.A.; Chalberg, T.W.; Schaffer, D.V. Viral Vectors for Gene Therapy: Translational and Clinical Outlook. Annu. Rev. Biomed. Eng. 2015, 17, 63–89.
- Xu, C.L.; Ruan, M.Z.C.; Mahajan, V.B.; Tsang, S.H. Viral Delivery Systems for CRISPR. Viruses 2019, 11, 28.
- Wan, T.; Niu, D.; Wu, C.; Xu, F.-J.; Church, G.; Ping, Y. Material Solutions for Delivery of CRISPR/Cas-Based Genome Editing Tools: Current Status and Future Outlook. Mater. Today 2019, 26, 40–66.
- Navarro-Serna, S.; Vilarino, M.; Park, I.; Gadea, J.; Ross, P.J. Livestock Gene Editing by One-Step Embryo Manipulation. J. Equine Vet. Sci. 2020, 89, 103025.
- Bhandawat, A.; Sharma, V.; Rishi, V.; Roy, J.K. Biolistic Delivery of Programmable Nuclease (CRISPR/Cas9) in Bread Wheat. In Methods in Molecular Biology; Springer: New York, NY, USA, 2020; pp. 309–329. ISBN 9781071603550.
- Liang, Z.; Chen, K.; Gao, C. Biolistic Delivery of CRISPR/Cas9 with Ribonucleoprotein Complex in Wheat. In Methods in Molecular Biology; Springer: New York, NY, USA, 2019; pp. 327–335. ISBN 9781493989904.
- Seki, A.; Rutz, S. Optimized RNP Transfection for Highly Efficient CRISPR/Cas9-Mediated Gene Knockout in Primary T Cells. J. Exp. Med. 2018, 215, 985–997.
- Huang, R.-S.; Shih, H.-A.; Lai, M.-C.; Chang, Y.-J.; Lin, S. Enhanced NK-92 Cytotoxicity by CRISPR Genome Engineering Using Cas9 Ribonucleoproteins. Front. Immunol. 2020, 11, 1008.
- Kim, S.; Kim, D.; Cho, S.W.; Kim, J.; Kim, J.-S. Highly Efficient RNA-Guided Genome Editing in Human Cells via Delivery of Purified Cas9 Ribonucleoproteins. Genome Res. 2014, 24, 1012–1019.
- Modarai, S.R.; Man, D.; Bialk, P.; Rivera-Torres, N.; Bloh, K.; Kmiec, E.B. Efficient Delivery and Nuclear Uptake Is Not Sufficient to Detect Gene Editing in CD34+ Cells Directed by a Ribonucleoprotein Complex. Mol. Ther. Nucleic Acids 2018, 11, 116–129.
- Hiatt, J.; Cavero, D.A.; McGregor, M.J.; Zheng, W.; Budzik, J.M.; Roth, T.L.; Haas, K.M.; Wu, D.; Rathore, U.; Meyer-Franke, A.; et al. Efficient Generation of Isogenic Primary Human Myeloid Cells Using CRISPR-Cas9 Ribonucleoproteins. Cell Rep. 2021, 35, 109105.
- Han, X.; Liu, Z.; Ma, Y.; Zhang, K.; Qin, L. Cas9 Ribonucleoprotein Delivery via Microfluidic Cell-deformation Chip for Human T-cell Genome Editing and Immunotherapy. Adv. Biosyst. 2017, 1, e1600007.
- Jarrell, J.A.; Sytsma, B.J.; Wilson, L.H.; Pan, F.L.; Lau, K.H.W.J.; Kirby, G.T.S.; Lievano, A.A.; Pawell, R.S. Genome Editing Human Primary T Cells with Microfluidic Vortex Shedding & CRISPR Cas9. bioRxiv 2020.
- Yen, J.; Fiorino, M.; Liu, Y.; Paula, S.; Clarkson, S.; Quinn, L.; Tschantz, W.R.; Klock, H.; Guo, N.; Russ, C.; et al. TRIAMF: A New Method for Delivery of Cas9 Ribonucleoprotein Complex to Human Hematopoietic Stem Cells. Sci. Rep. 2018, 8, 16304.
- Chen, Y.; Aslanoglou, S.; Murayama, T.; Gervinskas, G.; Fitzgerald, L.I.; Sriram, S.; Tian, J.; Johnston, A.P.R.; Morikawa, Y.; Suu, K.; et al. Silicon-nanotube-mediated Intracellular Delivery Enables Ex Vivo Gene Editing. Adv. Mater. 2020, 32, e2000036.
- Kholosy, W.M.; Visscher, M.; Ogink, K.; Buttstedt, H.; Griffin, K.; Beier, A.; Gerlach, J.P.; Molenaar, J.J.; Geijsen, N.; de Boer, M.; et al. Simple, Fast and Efficient ITOP-Mediated Delivery of CRISPR/Cas9 RNP in Difficult-to-Transduce Human Cells Including Primary T Cells. J. Biotechnol. 2021, 338, 71–80.
- Wei, T.; Cheng, Q.; Min, Y.-L.; Olson, E.N.; Siegwart, D.J. Systemic Nanoparticle Delivery of CRISPR-Cas9 Ribonucleoproteins for Effective Tissue Specific Genome Editing. Nat. Commun. 2020, 11, 3232.
- Gustafsson, O.; Rädler, J.; Roudi, S.; Lehto, T.; Hällbrink, M.; Lehto, T.; Gupta, D.; Andaloussi, S.E.L.; Nordin, J.Z. Efficient Peptide-Mediated in Vitro Delivery of Cas9 RNP. Pharmaceutics 2021, 13, 878.
- Zhang, S.; Shen, J.; Li, D.; Cheng, Y. Strategies in the Delivery of Cas9 Ribonucleoprotein for CRISPR/Cas9 Genome Editing. Theranostics 2021, 11, 614–648.
- Liu, C.; Wan, T.; Wang, H.; Zhang, S.; Ping, Y.; Cheng, Y. A Boronic Acid–Rich Dendrimer with Robust and Unprecedented Efficiency for Cytosolic Protein Delivery and CRISPR-Cas9 Gene Editing. Sci. Adv. 2019, 5, eaaw8922.
- Qiao, J.; Sun, W.; Lin, S.; Jin, R.; Ma, L.; Liu, Y. Cytosolic Delivery of CRISPR/Cas9 Ribonucleoproteins for Genome Editing Using Chitosan-Coated Red Fluorescent Protein. Chem. Commun. 2019, 55, 4707–4710.
- Ding, F.; Huang, X.; Gao, X.; Xie, M.; Pan, G.; Li, Q.; Song, J.; Zhu, X.; Zhang, C. A Non-Cationic Nucleic Acid Nanogel for the Delivery of the CRISPR/Cas9 Gene Editing Tool. Nanoscale 2019, 11, 17211–17215.
- Shahbazi, R.; Sghia-Hughes, G.; Reid, J.L.; Kubek, S.; Haworth, K.G.; Humbert, O.; Kiem, H.-P.; Adair, J.E. Targeted Homology-Directed Repair in Blood Stem and Progenitor Cells with CRISPR Nanoformulations. Nat. Mater. 2019, 18, 1124–1132.
- Alyami, M.Z.; Alsaiari, S.K.; Li, Y.; Qutub, S.S.; Aleisa, F.A.; Sougrat, R.; Merzaban, J.S.; Khashab, N.M. Cell-Type-Specific CRISPR/Cas9 Delivery by Biomimetic Metal Organic Frameworks. J. Am. Chem. Soc. 2020, 142, 1715–1720.
- Yue, H.; Zhou, X.; Cheng, M.; Xing, D. Graphene Oxide-Mediated Cas9/SgRNA Delivery for Efficient Genome Editing. Nanoscale 2018, 10, 1063–1071.
- Zhou, W.; Cui, H.; Ying, L.; Yu, X.-F. Enhanced Cytosolic Delivery and Release of CRISPR/Cas9 by Black Phosphorus Nanosheets for Genome Editing. Angew. Chem. Int. Ed. Engl. 2018, 57, 10268–10272.
- Li, S.; Song, Z.; Liu, C.; Chen, X.-L.; Han, H. Biomimetic Mineralization-Based CRISPR/Cas9 Ribonucleoprotein Nanoparticles for Gene Editing. ACS Appl. Mater. Interfaces 2019, 11, 47762–47770.
- Tyumentseva, M.A.; Tyumentsev, A.I.; Akimkin, V.G. Protocol for Assessment of the Efficiency of CRISPR/Cas RNP Delivery to Different Types of Target Cells. PLoS ONE 2021, 16, e0259812.
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic Acid Detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442.
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a Target Binding Unleashes Indiscriminate Single-Stranded DNase Activity. Science 2018, 360, 436–439.
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and Portable Nucleic Acid Detection Platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444.
- Myhrvold, C.; Freije, C.A.; Gootenberg, J.S.; Abudayyeh, O.O.; Metsky, H.C.; Durbin, A.F.; Kellner, M.J.; Tan, A.L.; Paul, L.M.; Parham, L.A.; et al. Field-Deployable Viral Diagnostics Using CRISPR-Cas13. Science 2018, 360, 444–448.
- Pardee, K.; Green, A.A.; Takahashi, M.K.; Braff, D.; Lambert, G.; Lee, J.W.; Ferrante, T.; Ma, D.; Donghia, N.; Fan, M.; et al. Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell 2016, 165, 1255–1266.
- Müller, V.; Rajer, F.; Frykholm, K.; Nyberg, L.K.; Quaderi, S.; Fritzsche, J.; Kristiansson, E.; Ambjörnsson, T.; Sandegren, L.; Westerlund, F. Direct Identification of Antibiotic Resistance Genes on Single Plasmid Molecules Using CRISPR/Cas9 in Combination with Optical DNA Mapping. Sci. Rep. 2016, 6, 37938.
- Zhou, W.; Hu, L.; Ying, L.; Zhao, Z.; Chu, P.K.; Yu, X.-F. A CRISPR–Cas9-Triggered Strand Displacement Amplification Method for Ultrasensitive DNA Detection. Nat. Commun. 2018, 9, 5012.
- Huang, M.; Zhou, X.; Wang, H.; Xing, D. Clustered Regularly Interspaced Short Palindromic Repeats/Cas9 Triggered Isothermal Amplification for Site-Specific Nucleic Acid Detection. Anal. Chem. 2018, 90, 2193–2200.
- Hajian, R.; Balderston, S.; Tran, T.; deBoer, T.; Etienne, J.; Sandhu, M.; Wauford, N.A.; Chung, J.-Y.; Nokes, J.; Athaiya, M.; et al. Detection of Unamplified Target Genes via CRISPR–Cas9 Immobilized on a Graphene Field-Effect Transistor. Nat. Biomed. Eng. 2019, 3, 427–437.
- Quan, J.; Langelier, C.; Kuchta, A.; Batson, J.; Teyssier, N.; Lyden, A.; Caldera, S.; McGeever, A.; Dimitrov, B.; King, R.; et al. FLASH: A next-Generation CRISPR Diagnostic for Multiplexed Detection of Antimicrobial Resistance Sequences. Nucleic Acids Res. 2019, 47, e83.
- Azhar, M.; Phutela, R.; Kumar, M.; Ansari, A.H.; Rauthan, R.; Gulati, S.; Sharma, N.; Sinha, D.; Sharma, S.; Singh, S.; et al. Rapid and Accurate Nucleobase Detection Using FnCas9 and Its Application in COVID-19 Diagnosis. Biosens. Bioelectron. 2021, 183, 113207.
- Wang, X.; Xiong, E.; Tian, T.; Cheng, M.; Lin, W.; Sun, J.; Zhou, X. CASLFA: CRISPR/Cas9-Mediated Lateral Flow Nucleic Acid Assay. bioRxiv 2019, 702209.
- Marsic, T.; Ali, Z.; Tehseen, M.; Mahas, A.; Hamdan, S.; Mahfouz, M. Vigilant: An Engineered VirD2-Cas9 Complex for Lateral Flow Assay-Based Detection of SARS-CoV2. Nano Lett. 2021, 21, 3596–3603.
- Osborn, M.J.; Bhardwaj, A.; Bingea, S.P.; Knipping, F.; Feser, C.J.; Lees, C.J.; Collins, D.P.; Steer, C.J.; Blazar, B.R.; Tolar, J. CRISPR/Cas9-Based Lateral Flow and Fluorescence Diagnostics. Bioengineering 2021, 8, 23.
- Jiao, C.; Sharma, S.; Dugar, G.; Peeck, N.L.; Bischler, T.; Wimmer, F.; Yu, Y.; Barquist, L.; Schoen, C.; Kurzai, O.; et al. Noncanonical CrRNAs Derived from Host Transcripts Enable Multiplexable RNA Detection by Cas9. Science 2021, 372, 941–948.
- Ali, Z.; Sánchez, E.; Tehseen, M.; Mahas, A.; Marsic, T.; Aman, R.; Sivakrishna Rao, G.; Alhamlan, F.S.; Alsanea, M.S.; Al-Qahtani, A.A.; et al. Bio-SCAN: A CRISPR/DCas9-Based Lateral Flow Assay for Rapid, Specific, and Sensitive Detection of SARS-CoV-2. ACS Synth. Biol. 2022, 11, 406–419.
- Wu, S.-S.; Li, Q.-C.; Yin, C.-Q.; Xue, W.; Song, C.-Q. Advances in CRISPR/Cas-Based Gene Therapy in Human Genetic Diseases. Theranostics 2020, 10, 4374–4382.
- Papasavva, P.; Kleanthous, M.; Lederer, C.W. Rare Opportunities: CRISPR/Cas-Based Therapy Development for Rare Genetic Diseases. Mol. Diagn. Ther. 2019, 23, 201–222.
- Kennedy, E.M.; Cullen, B.R. Gene Editing: A New Tool for Viral Disease. Annu. Rev. Med. 2017, 68, 401–411.
- Xiong, X.; Chen, M.; Lim, W.A.; Zhao, D.; Qi, L.S. CRISPR/Cas9 for Human Genome Engineering and Disease Research. Annu. Rev. Genom. Hum. Genet. 2016, 17, 131–154.
- Ferdosi, S.R.; Ewaisha, R.; Moghadam, F.; Krishna, S.; Park, J.G.; Ebrahimkhani, M.R.; Kiani, S.; Anderson, K.S. Multifunctional CRISPR-Cas9 with Engineered Immunosilenced Human T Cell Epitopes. Nat. Commun. 2019, 10, 1842.
- Doms, R.W. Chemokine Receptors and HIV Entry. AIDS 2001, 15, S34–S35.
- Koujah, L.; Shukla, D.; Naqvi, A.R. CRISPR-Cas Based Targeting of Host and Viral Genes as an Antiviral Strategy. Semin. Cell Dev. Biol. 2019, 96, 53–64.
- Liu, Z.; Chen, S.; Jin, X.; Wang, Q.; Yang, K.; Li, C.; Xiao, Q.; Hou, P.; Liu, S.; Wu, S.; et al. Genome Editing of the HIV Co-Receptors CCR5 and CXCR4 by CRISPR-Cas9 Protects CD4+ T Cells from HIV-1 Infection. Cell Biosci. 2017, 7, 47.
- Xu, L.; Yang, H.; Gao, Y.; Chen, Z.; Xie, L.; Liu, Y.; Liu, Y.; Wang, X.; Li, H.; Lai, W.; et al. CRISPR/Cas9-Mediated CCR5 Ablation in Human Hematopoietic Stem/Progenitor Cells Confers HIV-1 Resistance in Vivo. Mol. Ther. 2017, 25, 1782–1789.
- Li, C.; Guan, X.; Du, T.; Jin, W.; Wu, B.; Liu, Y.; Wang, P.; Hu, B.; Griffin, G.E.; Shattock, R.J.; et al. Inhibition of HIV-1 Infection of Primary CD4+ T-Cells by Gene Editing of CCR5 Using Adenovirus-Delivered CRISPR/Cas9. J. Gen. Virol. 2015, 96, 2381–2393.
- Cho, S.W.; Kim, S.; Kim, Y.; Kweon, J.; Kim, H.S.; Bae, S.; Kim, J.-S. Analysis of Off-Target Effects of CRISPR/Cas-Derived RNA-Guided Endonucleases and Nickases. Genome Res. 2014, 24, 132–141.
- Hou, P.; Chen, S.; Wang, S.; Yu, X.; Chen, Y.; Jiang, M.; Zhuang, K.; Ho, W.; Hou, W.; Huang, J.; et al. Genome Editing of CXCR4 by CRISPR/Cas9 Confers Cells Resistant to HIV-1 Infection. Sci. Rep. 2015, 5, 15577.
- Schumann, K.; Lin, S.; Boyer, E.; Simeonov, D.R.; Subramaniam, M.; Gate, R.E.; Haliburton, G.E.; Ye, C.J.; Bluestone, J.A.; Doudna, J.A.; et al. Generation of Knock-in Primary Human T Cells Using Cas9 Ribonucleoproteins. Proc. Natl. Acad. Sci. USA 2015, 112, 10437–10442.
- Liu, S.; Wang, Q.; Yu, X.; Li, Y.; Guo, Y.; Liu, Z.; Sun, F.; Hou, W.; Li, C.; Wu, L.; et al. HIV-1 Inhibition in Cells with CXCR4 Mutant Genome Created by CRISPR-Cas9 and PiggyBac Recombinant Technologies. Sci. Rep. 2018, 8, 8573.
- Strich, J.R.; Chertow, D.S. CRISPR-Cas Biology and Its Application to Infectious Diseases. J. Clin. Microbiol. 2019, 57, e01307-18.
- Hu, W.; Kaminski, R.; Yang, F.; Zhang, Y.; Cosentino, L.; Li, F.; Luo, B.; Alvarez-Carbonell, D.; Garcia-Mesa, Y.; Karn, J.; et al. RNA-Directed Gene Editing Specifically Eradicates Latent and Prevents New HIV-1 Infection. Proc. Natl. Acad. Sci. USA 2014, 111, 11461–11466.
- Wang, G.; Zhao, N.; Berkhout, B.; Das, A.T. CRISPR-Cas Based Antiviral Strategies against HIV-1. Virus Res. 2018, 244, 321–332.
- Yin, C.; Zhang, T.; Qu, X.; Zhang, Y.; Putatunda, R.; Xiao, X.; Li, F.; Xiao, W.; Zhao, H.; Dai, S.; et al. In Vivo Excision of HIV-1 Provirus by SaCas9 and Multiplex Single-Guide RNAs in Animal Models. Mol. Ther. 2017, 25, 1168–1186.
- Bella, R.; Kaminski, R.; Mancuso, P.; Young, W.-B.; Chen, C.; Sariyer, R.; Fischer, T.; Amini, S.; Ferrante, P.; Jacobson, J.M.; et al. Removal of HIV DNA by CRISPR from Patient Blood Engrafts in Humanized Mice. Mol. Ther. Nucleic Acids 2018, 12, 275–282.
- Li, H.; Sheng, C.; Wang, S.; Yang, L.; Liang, Y.; Huang, Y.; Liu, H.; Li, P.; Yang, C.; Yang, X.; et al. Removal of Integrated Hepatitis B Virus DNA Using CRISPR-Cas9. Front. Cell. Infect. Microbiol. 2017, 7, 91.
- Scott, T.; Moyo, B.; Nicholson, S.; Maepa, M.B.; Watashi, K.; Ely, A.; Weinberg, M.S.; Arbuthnot, P. SsAAVs Containing Cassettes Encoding SaCas9 and Guides Targeting Hepatitis B Virus Inactivate Replication of the Virus in Cultured Cells. Sci. Rep. 2017, 7, 7401.
- van Diemen, F.R.; Kruse, E.M.; Hooykaas, M.J.G.; Bruggeling, C.E.; Schürch, A.C.; van Ham, P.M.; Imhof, S.M.; Nijhuis, M.; Wiertz, E.J.H.J.; Lebbink, R.J. CRISPR/Cas9-Mediated Genome Editing of Herpesviruses Limits Productive and Latent Infections. PLoS Pathog. 2016, 12, e1005701.
- Roehm, P.C.; Shekarabi, M.; Wollebo, H.S.; Bellizzi, A.; He, L.; Salkind, J.; Khalili, K. Inhibition of HSV-1 Replication by Gene Editing Strategy. Sci. Rep. 2016, 6, 23146.
- Wollebo, H.S.; Bellizzi, A.; Kaminski, R.; Hu, W.; White, M.K.; Khalili, K. CRISPR/Cas9 System as an Agent for Eliminating Polyomavirus JC Infection. PLoS ONE 2015, 10, e0136046.
- Kennedy, E.M.; Kornepati, A.V.R.; Goldstein, M.; Bogerd, H.P.; Poling, B.C.; Whisnant, A.W.; Kastan, M.B.; Cullen, B.R. Inactivation of the Human Papillomavirus E6 or E7 Gene in Cervical Carcinoma Cells by Using a Bacterial CRISPR/Cas RNA-Guided Endonuclease. J. Virol. 2014, 88, 11965–11972.
- Vaidyanathan, S.; Salahudeen, A.A.; Sellers, Z.M.; Bravo, D.T.; Choi, S.S.; Batish, A.; Le, W.; Baik, R.; de la O, S.; Kaushik, M.P.; et al. High-Efficiency, Selection-Free Gene Repair in Airway Stem Cells from Cystic Fibrosis Patients Rescues CFTR Function in Differentiated Epithelia. Cell Stem Cell 2020, 26, 161–171.e4.
- Schwank, G.; Koo, B.-K.; Sasselli, V.; Dekkers, J.F.; Heo, I.; Demircan, T.; Sasaki, N.; Boymans, S.; Cuppen, E.; van der Ent, C.K.; et al. Functional Repair of CFTR by CRISPR/Cas9 in Intestinal Stem Cell Organoids of Cystic Fibrosis Patients. Cell Stem Cell 2013, 13, 653–658.
- Firth, A.L.; Menon, T.; Parker, G.S.; Qualls, S.J.; Lewis, B.M.; Ke, E.; Dargitz, C.T.; Wright, R.; Khanna, A.; Gage, F.H.; et al. Functional Gene Correction for Cystic Fibrosis in Lung Epithelial Cells Generated from Patient IPSCs. Cell Rep. 2015, 12, 1385–1390.
- Wang, G. Genome Editing for Cystic Fibrosis. Cells 2023, 12, 1555.
- Vakulskas, C.A.; Dever, D.P.; Rettig, G.R.; Turk, R.; Jacobi, A.M.; Collingwood, M.A.; Bode, N.M.; McNeill, M.S.; Yan, S.; Camarena, J.; et al. A High-Fidelity Cas9 Mutant Delivered as a Ribonucleoprotein Complex Enables Efficient Gene Editing in Human Hematopoietic Stem and Progenitor Cells. Nat. Med. 2018, 24, 1216–1224.
- Lomova, A.; Clark, D.N.; Campo-Fernandez, B.; Flores-Bjurström, C.; Kaufman, M.L.; Fitz-Gibbon, S.; Wang, X.; Miyahira, E.Y.; Brown, D.; DeWitt, M.A.; et al. Improving Gene Editing Outcomes in Human Hematopoietic Stem and Progenitor Cells by Temporal Control of DNA Repair. Stem Cells 2019, 37, 284–294.
- Martin, R.M.; Ikeda, K.; Cromer, M.K.; Uchida, N.; Nishimura, T.; Romano, R.; Tong, A.J.; Lemgart, V.T.; Camarena, J.; Pavel-Dinu, M.; et al. Highly Efficient and Marker-Free Genome Editing of Human Pluripotent Stem Cells by CRISPR-Cas9 RNP and AAV6 Donor-Mediated Homologous Recombination. Cell Stem Cell 2019, 24, 821–828.e5.
- Park, S.H.; Bao, G. CRISPR/Cas9 Gene Editing for Curing Sickle Cell Disease. Transfus. Apher. Sci. 2021, 60, 103060.
- Wattanapanitch, M.; Damkham, N.; Potirat, P.; Trakarnsanga, K.; Janan, M.; U-pratya, Y.; Kheolamai, P.; Klincumhom, N.; Issaragrisil, S. One-Step Genetic Correction of Hemoglobin E/Beta-Thalassemia Patient-Derived IPSCs by the CRISPR/Cas9 System. Stem Cell Res. Ther. 2018, 9, 46.
- Patsali, P.; Turchiano, G.; Papasavva, P.; Romito, M.; Loucari, C.C.; Stephanou, C.; Christou, S.; Sitarou, M.; Mussolino, C.; Cornu, T.I.; et al. Correction of IVS I-110(G>A) β-Thalassemia by CRISPR/Cas-and TALEN-Mediated Disruption of Aberrant Regulatory Elements in Human Hematopoietic Stem and Progenitor Cells. Haematologica 2019, 104, e497–e501.
- Niu, X.; He, W.; Song, B.; Ou, Z.; Fan, D.; Chen, Y.; Fan, Y.; Sun, X. Combining Single Strand Oligodeoxynucleotides and CRISPR/Cas9 to Correct Gene Mutations in β-Thalassemia-Induced Pluripotent Stem Cells. J. Biol. Chem. 2016, 291, 16576–16585.
- Gamage, U.; Warnakulasuriya, K.; Hansika, S.; Silva, G.N. CRISPR Gene Therapy: A Promising One-Time Therapeutic Approach for Transfusion-Dependent β-Thalassemia—CRISPR-Cas9 Gene Editing for β-Thalassemia. Thalass. Rep. 2023, 13, 51–69.
- Monteys, A.M.; Ebanks, S.A.; Keiser, M.S.; Davidson, B.L. CRISPR/Cas9 Editing of the Mutant Huntingtin Allele in Vitro and in Vivo. Mol. Ther. 2017, 25, 12–23.
- Yang, S.; Chang, R.; Yang, H.; Zhao, T.; Hong, Y.; Kong, H.E.; Sun, X.; Qin, Z.; Jin, P.; Li, S.; et al. CRISPR/Cas9-Mediated Gene Editing Ameliorates Neurotoxicity in Mouse Model of Huntington’s Disease. J. Clin. Investig. 2017, 127, 2719–2724.
- Merienne, N.; Vachey, G.; de Longprez, L.; Meunier, C.; Zimmer, V.; Perriard, G.; Canales, M.; Mathias, A.; Herrgott, L.; Beltraminelli, T.; et al. The Self-Inactivating KamiCas9 System for the Editing of CNS Disease Genes. Cell Rep. 2017, 20, 2980–2991.
- Ekman, F.K.; Ojala, D.S.; Adil, M.M.; Lopez, P.A.; Schaffer, D.V.; Gaj, T. CRISPR-Cas9-Mediated Genome Editing Increases Lifespan and Improves Motor Deficits in a Huntington’s Disease Mouse Model. Mol. Ther. Nucleic Acids 2019, 17, 829–839.
- Alkanli, S.S.; Alkanli, N.; Ay, A.; Albeniz, I. CRISPR/Cas9 Mediated Therapeutic Approach in Huntington’s Disease. Mol. Neurobiol. 2023, 60, 1486–1498.
- Nelson, C.E.; Hakim, C.H.; Ousterout, D.G.; Thakore, P.I.; Moreb, E.A.; Rivera, R.M.C.; Madhavan, S.; Pan, X.; Ran, F.A.; Yan, W.X.; et al. In Vivo Genome Editing Improves Muscle Function in a Mouse Model of Duchenne Muscular Dystrophy. Science 2016, 351, 403–407.
- Nelson, C.E.; Wu, Y.; Gemberling, M.P.; Oliver, M.L.; Waller, M.A.; Bohning, J.D.; Robinson-Hamm, J.N.; Bulaklak, K.; Castellanos Rivera, R.M.; Collier, J.H.; et al. Long-Term Evaluation of AAV-CRISPR Genome Editing for Duchenne Muscular Dystrophy. Nat. Med. 2019, 25, 427–432.
- Min, Y.-L.; Li, H.; Rodriguez-Caycedo, C.; Mireault, A.A.; Huang, J.; Shelton, J.M.; McAnally, J.R.; Amoasii, L.; Mammen, P.P.A.; Bassel-Duby, R.; et al. CRISPR-Cas9 Corrects Duchenne Muscular Dystrophy Exon 44 Deletion Mutations in Mice and Human Cells. Sci. Adv. 2019, 5, eaav4324.
- Bengtsson, N.E.; Hall, J.K.; Odom, G.L.; Phelps, M.P.; Andrus, C.R.; Hawkins, R.D.; Hauschka, S.D.; Chamberlain, J.R.; Chamberlain, J.S. Muscle-Specific CRISPR/Cas9 Dystrophin Gene Editing Ameliorates Pathophysiology in a Mouse Model for Duchenne Muscular Dystrophy. Nat. Commun. 2017, 8, 14454.
- Amoasii, L.; Hildyard, J.C.W.; Li, H.; Sanchez-Ortiz, E.; Mireault, A.; Caballero, D.; Harron, R.; Stathopoulou, T.-R.; Massey, C.; Shelton, J.M.; et al. Gene Editing Restores Dystrophin Expression in a Canine Model of Duchenne Muscular Dystrophy. Science 2018, 362, 86–91.
- Zhang, Y.; Long, C.; Li, H.; McAnally, J.R.; Baskin, K.K.; Shelton, J.M.; Bassel-Duby, R.; Olson, E.N. CRISPR-Cpf1 Correction of Muscular Dystrophy Mutations in Human Cardiomyocytes and Mice. Sci. Adv. 2017, 3, e1602814.
- Agrawal, P.; Harish, V.; Mohd, S.; Singh, S.K.; Tewari, D.; Tatiparthi, R.; Harshita; Vishwas, S.; Sutrapu, S.; Dua, K.; et al. Role of CRISPR/Cas9 in the Treatment of Duchenne Muscular Dystrophy and Its Delivery Strategies. Life Sci. 2023, 330, 122003.
- Stephens, C.J.; Lauron, E.J.; Kashentseva, E.; Lu, Z.H.; Yokoyama, W.M.; Curiel, D.T. Long-Term Correction of Hemophilia B Using Adenoviral Delivery of CRISPR/Cas9. J. Control. Release 2019, 298, 128–141.
- Hu, Z.; Zhou, M.; Wu, Y.; Li, Z.; Liu, X.; Wu, L.; Liang, D. SsODN-Mediated in-Frame Deletion with CRISPR/Cas9 Restores FVIII Function in Hemophilia A-Patient-Derived IPSCs and ECs. Mol. Ther. Nucleic Acids 2019, 17, 198–209.
- Lyu, C.; Shen, J.; Wang, R.; Gu, H.; Zhang, J.; Xue, F.; Liu, X.; Liu, W.; Fu, R.; Zhang, L.; et al. Targeted Genome Engineering in Human Induced Pluripotent Stem Cells from Patients with Hemophilia B Using the CRISPR-Cas9 System. Stem Cell Res. Ther. 2018, 9, 92.
- Chen, H.; Shi, M.; Gilam, A.; Zheng, Q.; Zhang, Y.; Afrikanova, I.; Li, J.; Gluzman, Z.; Jiang, R.; Kong, L.-J.; et al. Hemophilia A Ameliorated in Mice by CRISPR-Based in Vivo Genome Editing of Human Factor VIII. Sci. Rep. 2019, 9, 16838.
- Hiramoto, T.; Kashiwakura, Y.; Hayakawa, M.; Baatartsogt, N.; Kamoshita, N.; Abe, T.; Inaba, H.; Nishimasu, H.; Uosaki, H.; Hanazono, Y.; et al. PAM-Flexible Cas9-Mediated Base Editing of a Hemophilia B Mutation in Induced Pluripotent Stem Cells. Commun. Med. 2023, 3, 56.
- Chung, J.Y.; Ain, Q.U.; Song, Y.; Yong, S.-B.; Kim, Y.-H. Targeted Delivery of CRISPR Interference System against Fabp4 to White Adipocytes Ameliorates Obesity, Inflammation, Hepatic Steatosis, and Insulin Resistance. Genome Res. 2019, 29, 1442–1452.
- Maxwell, K.G.; Augsornworawat, P.; Velazco-Cruz, L.; Kim, M.H.; Asada, R.; Hogrebe, N.J.; Morikawa, S.; Urano, F.; Millman, J.R. Gene-Edited Human Stem Cell–Derived β Cells from a Patient with Monogenic Diabetes Reverse Preexisting Diabetes in Mice. Sci. Transl. Med. 2020, 12, eaax9106.
- Cho, E.Y.; Ryu, J.-Y.; Lee, H.A.R.; Hong, S.H.; Park, H.S.; Hong, K.S.; Park, S.-G.; Kim, H.P.; Yoon, T.-J. Lecithin Nano-Liposomal Particle as a CRISPR/Cas9 Complex Delivery System for Treating Type 2 Diabetes. J. Nanobiotechnol. 2019, 17, 19.
- Grotz, A.K.; Abaitua, F.; Navarro-Guerrero, E.; Hastoy, B.; Ebner, D.; Gloyn, A.L. A CRISPR/Cas9 Genome Editing Pipeline in the EndoC-ΒH1 Cell Line to Study Genes Implicated in Beta Cell Function. Wellcome Open Res. 2020, 4, 150.
- Cheng, Y.; Wang, H.; Li, M. The Promise of CRISPR/Cas9 Technology in Diabetes Mellitus Therapy: How Gene Editing Is Revolutionizing Diabetes Research and Treatment. J. Diabetes Complicat. 2023, 37, 108524.
- Olivaes, J.; Bonamino, M.H.; Markoski, M.M. CRISPR/Cas 9 System for the Treatment of Dilated Cardiomyopathy: A Hypothesis Related to Function of a MAP Kinase. Med. Hypotheses 2019, 128, 91–93.
- Zhao, H.; Li, Y.; He, L.; Pu, W.; Yu, W.; Li, Y.; Wu, Y.-T.; Xu, C.; Wei, Y.; Ding, Q.; et al. In Vivo AAV-CRISPR/Cas9–Mediated Gene Editing Ameliorates Atherosclerosis in Familial Hypercholesterolemia. Circulation 2020, 141, 67–79.
- Caron, J.; Pène, V.; Tolosa, L.; Villaret, M.; Luce, E.; Fourrier, A.; Heslan, J.-M.; Saheb, S.; Bruckert, E.; Gómez-Lechón, M.J.; et al. Low-Density Lipoprotein Receptor-Deficient Hepatocytes Differentiated from Induced Pluripotent Stem Cells Allow Familial Hypercholesterolemia Modeling, CRISPR/Cas-Mediated Genetic Correction, and Productive Hepatitis C Virus Infection. Stem Cell Res. Ther. 2019, 10, 221.
- Wang, X.; Raghavan, A.; Chen, T.; Qiao, L.; Zhang, Y.; Ding, Q.; Musunuru, K. CRISPR-Cas9 Targeting of PCSK9 in Human Hepatocytes in Vivo—Brief Report. Arterioscler. Thromb. Vasc. Biol. 2016, 36, 783–786.
- Musunuru, K. CRISPR and Cardiovascular Diseases. Cardiovasc. Res. 2023, 119, 79–93.
- Liu, X.; Zhang, Y.; Cheng, C.; Cheng, A.W.; Zhang, X.; Li, N.; Xia, C.; Wei, X.; Liu, X.; Wang, H. CRISPR-Cas9-Mediated Multiplex Gene Editing in CAR-T Cells. Cell Res. 2017, 27, 154–157.
- Choi, B.D.; Yu, X.; Castano, A.P.; Darr, H.; Henderson, D.B.; Bouffard, A.A.; Larson, R.C.; Scarfò, I.; Bailey, S.R.; Gerhard, G.M.; et al. CRISPR-Cas9 Disruption of PD-1 Enhances Activity of Universal EGFRvIII CAR T Cells in a Preclinical Model of Human Glioblastoma. J. Immunother. Cancer 2019, 7, 304.
- McGowan, E.; Lin, Q.; Ma, G.; Yin, H.; Chen, S.; Lin, Y. PD-1 Disrupted CAR-T Cells in the Treatment of Solid Tumors: Promises and Challenges. Biomed. Pharmacother. 2020, 121, 109625.
- Guo, X.; Jiang, H.; Shi, B.; Zhou, M.; Zhang, H.; Shi, Z.; Du, G.; Luo, H.; Wu, X.; Wang, Y.; et al. Disruption of PD-1 Enhanced the Anti-Tumor Activity of Chimeric Antigen Receptor T Cells against Hepatocellular Carcinoma. Front. Pharmacol. 2018, 9, 1118.
- Liu, J.; Zhou, G.; Zhang, L.; Zhao, Q. Building Potent Chimeric Antigen Receptor T Cells with CRISPR Genome Editing. Front. Immunol. 2019, 10, 456.
- Nakazawa, T.; Natsume, A.; Nishimura, F.; Morimoto, T.; Matsuda, R.; Nakamura, M.; Yamada, S.; Nakagawa, I.; Motoyama, Y.; Park, Y.-S.; et al. Effect of CRISPR/Cas9-Mediated PD-1-Disrupted Primary Human Third-Generation CAR-T Cells Targeting EGFRvIII on in Vitro Human Glioblastoma Cell Growth. Cells 2020, 9, 998.
- Ren, J.; Zhang, X.; Liu, X.; Fang, C.; Jiang, S.; June, C.H.; Zhao, Y. A Versatile System for Rapid Multiplex Genome-Edited CAR T Cell Generation. Oncotarget 2017, 8, 17002–17011.
- Ren, J.; Liu, X.; Fang, C.; Jiang, S.; June, C.H.; Zhao, Y. Multiplex Genome Editing to Generate Universal CAR T Cells Resistant to PD1 Inhibition. Clin. Cancer Res. 2017, 23, 2255–2266.
- Zhang, Y.; Zhang, X.; Cheng, C.; Mu, W.; Liu, X.; Li, N.; Wei, X.; Liu, X.; Xia, C.; Wang, H. CRISPR-Cas9 Mediated LAG-3 Disruption in CAR-T Cells. Front. Med. 2017, 11, 554–562.
- Blaeschke, F.; Willier, S.; Stenger, D.; Lepenies, M.; Horstmann, M.A.; Escherich, G.; Zimmermann, M.; Rojas Ringeling, F.; Canzar, S.; Kaeuferle, T.; et al. Leukemia-Induced Dysfunctional TIM-3+CD4+ Bone Marrow T Cells Increase Risk of Relapse in Pediatric B-Precursor ALL Patients. Leukemia 2020, 34, 2607–2620.
- Stadtmauer, E.A.; Fraietta, J.A.; Davis, M.M.; Cohen, A.D.; Weber, K.L.; Lancaster, E.; Mangan, P.A.; Kulikovskaya, I.; Gupta, M.; Chen, F.; et al. CRISPR-Engineered T Cells in Patients with Refractory Cancer. Science 2020, 367, eaba7365.
- Wei, W.; Chen, Z.-N.; Wang, K. CRISPR/Cas9: A Powerful Strategy to Improve CAR-T Cell Persistence. Int. J. Mol. Sci. 2023, 24, 12317.
- Dimitri, A.; Herbst, F.; Fraietta, J.A. Engineering the Next-Generation of CAR T-Cells with CRISPR-Cas9 Gene Editing. Mol. Cancer 2022, 21, 78.
- Eyquem, J.; Mansilla-Soto, J.; Giavridis, T.; van der Stegen, S.J.C.; Hamieh, M.; Cunanan, K.M.; Odak, A.; Gönen, M.; Sadelain, M. Targeting a CAR to the TRAC Locus with CRISPR/Cas9 Enhances Tumour Rejection. Nature 2017, 543, 113–117.
- MacLeod, D.T.; Antony, J.; Martin, A.J.; Moser, R.J.; Hekele, A.; Wetzel, K.J.; Brown, A.E.; Triggiano, M.A.; Hux, J.A.; Pham, C.D.; et al. Integration of a CD19 CAR into the TCR Alpha Chain Locus Streamlines Production of Allogeneic Gene-Edited CAR T Cells. Mol. Ther. 2017, 25, 949–961.
- Liu, X.; Zhao, Y. CRISPR/Cas9 Genome Editing: Fueling the Revolution in Cancer Immunotherapy. Curr. Res. Transl. Med. 2018, 66, 39–42.
- Roth, T.L.; Puig-Saus, C.; Yu, R.; Shifrut, E.; Carnevale, J.; Li, P.J.; Hiatt, J.; Saco, J.; Krystofinski, P.; Li, H.; et al. Reprogramming Human T Cell Function and Specificity with Non-Viral Genome Targeting. Nature 2018, 559, 405–409.
