Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Brain tumor segmentation in medical imaging is a critical task for diagnosis and treatment while preserving patient data privacy and security. Traditional centralized approaches often encounter obstacles in data sharing due to privacy regulations and security concerns, hindering the development of advanced AI-based medical imaging applications.
- brain tumor segmentation
- computation cost
- data leakage
- data privacy
- deep learning
- federated learning
- medical imaging
1. Introduction
Brain tumors are irregular growths of cells developing within the brain. These tumors can be benign or malignant and can cause severe health complications including neurological deficits and death [1]. It is very important to correctly identify and define the boundaries of brain tumors so that doctors can diagnose them accurately, plan effective treatment strategies, and keep track of how effective the treatments are [2]. To obtain a better view of brain tumors and the structures surrounding them, medical professionals often rely on imaging methods like magnetic resonance imaging (MRI) and computed tomography (CT) scans. These techniques are regularly employed to help diagnose brain tumors and figure out the most effective treatment plan for the patient [3]. To better understand MRI and CT scans, these are divided into distinct segments, which is a practice known as brain tumor segmentation. This segmentation process corresponds to different parts of a tumor, including its necrotic core, the edema, and the enhancing tumor. This segmentation is crucial for accurate diagnosis and treatment planning as it involves the process of separating unhealthy tissues from healthy ones [4]. For many years, manual segmentation has been practiced by radiotherapists to perform these procedures. However, there was a high chance of the variability in the results between different observers and even in same observer. The process is time-consuming and can be affected by the expertise of the radiologist [5]. To overcome these limitations, automated and semi-automated segmentation algorithms have been developed.
The accurate and precise outlining of brain tumors in medical images is important for several reasons. It is a crucial part of diagnosing and determining the stage of the tumor. It helps doctors to understand how big the tumor is, where it is located, and how much it is affecting nearby tissues [6]. This information is critical for determining the prognosis and guiding personalized treatment strategies such as surgery, radiotherapy, and chemotherapy [7]. Accurate segmentation enables an unbiased and measurable evaluation of how tumors respond to treatment, reducing the subjective factors associated with manual evaluation [8]. This can help clinicians to make informed decisions about the efficacy of a particular treatment, and whether adjustments to the therapeutic plan are necessary. Precise segmentation can facilitate the development of computational models that predict tumor growth and its reaction to therapy, thus enhancing personalized medicine [9]. Such models can help to identify potential treatment targets, optimize treatment schedules, and identify patients at risk of tumor recurrence or progression. The increasing availability of medical imaging data and advancement in the technology has resulted in the development of robust and accurate computational segmentation algorithms [10]. However, the need for large amounts of annotated data brings its own challenges, i.e., the security and privacy concerns that limit the access to such data [11]. Additionally, in the context of deep learning models, the key challenge is the centralized nature of these approaches, which requires data from multiple institutions to be combined in a central repository. However, this process can present significant logistical difficulties and may also face limitations due to regulatory constraints.
To overcome the challenge of the centralized nature of deep learning (DL) and data security, the idea of federated learning (FL) is introduced that works via decentralized processing units which have their own data and model [12]. In FL, a unit does not share its data with other units, but instead, it shares the weights of the model with the centralized controller for the refinement of the overall model’s performance. Figure 1a,b show the working flow of federated learning. Such privacy and security assurance resulted in the popularity of FL in areas with significant data privacy concerns such as healthcare and medical imaging [13]. Since its inception, FL has largely been applied to such tasks, i.e., segmentation, reconstruction, and classification, with significantly reliable performance and results [14][15].
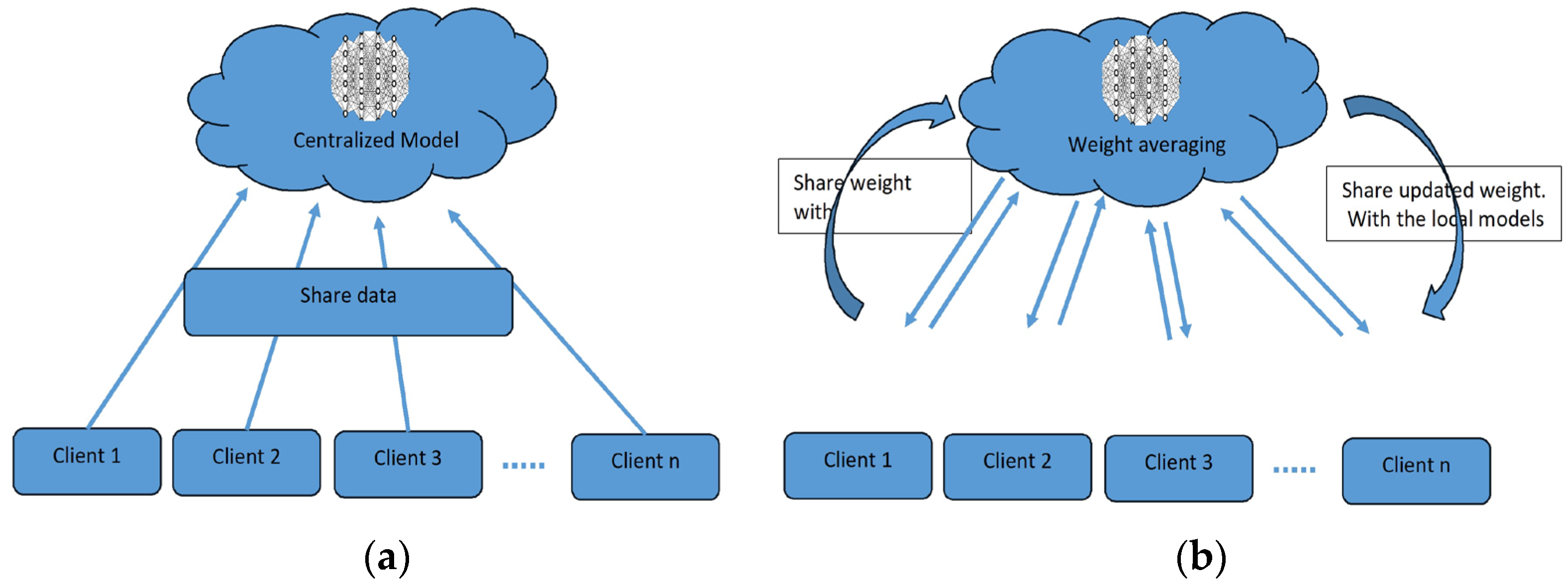
Figure 1. (a) Centralized model of FL; (b) client server model of FL.
In brain tumor segmentation, FL can utilize data from multiple institutions, enhancing the performance and generalizability of segmentation algorithms. This is accomplished while maintaining the utmost data privacy and security. FL is crucial in addressing the heterogeneity of the brain tumor, which varies in location, shape, size, and intensity across patients. By leveraging data from multiple institutions, FL improves segmentation algorithms while preserving data privacy and security [16]. By allowing institutions to collaborate without sharing raw data, FL can enable the development of more robust and accurate segmentation algorithms that can better account for this variability.
2. Traditional Brain Tumor Segmentation
Several conventional techniques have been proposed for brain tumor segmentation, including region-focused, edge-focused, and model-focused approaches [17]. While conventional brain tumor segmentation techniques have been widely used in medical imaging, they have several limitations, including sensitivity to image noise, lack of robustness to variations in image intensity and texture, and dependence on expert knowledge for parameter tuning.
The accuracy of conventional techniques depends on the specific application and available resources [18]. Thus, the choice of the most appropriate segmentation technique depends on various factors, including the image quality, the type and location of the tumor, and the clinical objectives. In recent years, deep-learning-based methods, such as CNNs, RNNs, and their variants, have emerged as powerful alternatives to conventional techniques, achieving state-of-the-art performance on various medical imaging tasks, including brain tumor segmentation. The accuracy of these techniques depends on the specific application and available resources. However, these methods require large and diverse datasets for training and the careful selection of appropriate network architectures and hyperparameters.
2.1. Region-Focused Approaches
Region-focused approaches have been widely used in brain tumor segmentation due to their simplicity and computational efficiency. These methods aim to segment brain tumors based on the intensity or texture differences between the tumor and its surrounding tissue. Several region-based approaches have been proposed for brain tumor segmentation, including thresholding, clustering, and region growing methods [19].
Thresholding-based methods involve setting a fixed threshold value to separate the tumor and non-tumor regions [20]. These methods are simple and fast, but may be affected by image noise and variations in image intensity and texture. Clustering-based methods use statistical clustering algorithms, such as fuzzy C-means, k-means, and expectation-maximization algorithms, to partition the image into tumor and non-tumor regions. Clustering-based methods can achieve better segmentation accuracy than thresholding-based methods, but they require the careful selection of clustering parameters.
Region growing methods start with a seed point within the tumor region and expand the region by incorporating adjacent pixels that meet specific criteria. Region growing methods can produce accurate and smooth segmentation results, but may be affected by variations in image intensity and texture, and they are sensitive to the selection of seed points. Several studies have compared the performance of region-based approaches for brain tumor segmentation. Jaglan et al. [21] compared the performance of various thresholding- and clustering-based methods and found that the Otsu thresholding method and the fuzzy C-means clustering method achieved the highest segmentation accuracy. Charutha et al. [22] compared the performance of various region growing methods and found that a fast-marching method with an adaptive threshold achieved the best segmentation results.
Region-focused approaches have been widely used in brain tumor segmentation due to their simplicity and computational efficiency. While these methods have their strengths, they may be negatively affected by variations in image intensity and texture. Thus, they require the careful selection of parameters and domain knowledge. Advances in deep-learning-based methods have shown promising results in overcoming some of the limitations of region-focused approaches.
2.2. Edge-Focused Approaches
Edge-focused approaches aim to identify the boundary between the tumor and surrounding tissue by detecting edges in the image. These methods often use edge detection techniques, such as Canny or Sobel filters, to highlight the edges and then apply a segmentation algorithm to separate the tumor from the background. Edge-focused approaches can achieve better segmentation accuracy than region-based approaches, but may be sensitive to image noise and produce fragmented results. Rajan and Sundar [23] combined K-means clustering, fuzzy C-means, and active contour techniques to enhance segmentation. Similarly, Sheela and Suganthi [24] used an approach to improve accuracy, reduce processing time, and compute tumor volume for effective diagnosis and treatment. The process involves collecting images, calculating tumor area and volume, preprocessing and enhancing the images, adjusting the image intensities, clustering the images, classifying the images, extracting features, and segmenting the images. Each step in the process is important and contributes to the overall quality of the enhanced images. The process has strengths such as its ability to improve the quality of MRI images in several ways, i.e., identifying tumors, providing quantitative information about tumors, and segmenting tumors from the surrounding tissue. However, the process also has weaknesses, such as its complexity, time-consuming nature, reliance on specialized software and hardware, difficulty in automation, and sensitivity to noise in the images.
Several studies have compared the performance of edge-focused approaches for brain tumor segmentation. The performance of various edge detection methods, including Canny, Sobel, and Laplacian filters, showed that the Sobel filter achieved the highest segmentation accuracy. Und et al. [25] compared the performance of edge detection methods combined with different segmentation algorithms, such as region growing and active contour methods, and found that the Sobel filter combined with the active contour method achieved the best segmentation results.
Edge-focused approaches can be computationally intensive due to the need to detect edges in the image. Moreover, it may be affected by variations in image intensity and texture and produce fragmented segmentation results. Deep-learning-based methods, such as CNNs, have shown promising results in overcoming some of these limitations. They can also achieve better segmentation accuracy than region-based approaches, but they may be sensitive to image noise and produce fragmented results. The choice of the most appropriate edge detection method depends on the specific application and available resources. Deep-learning-based techniques have reported very significant results in improving segmentation accuracy and robustness.
2.3. Model-Focused Approaches
Model-focused approaches aim to model the shape, texture, and intensity-based characteristics of brain tumors and their surrounding tissue to improve segmentation accuracy. These methods often use traditional machine learning algorithms, such as random forests, decision trees, and support vector machines, to learn those features that distinguish between tumorous and non-tumorous regions [23]. Model-focused approaches can achieve high segmentation accuracy and robustness subject to the availability of high-quality large datasets for training and may be affected by overfitting, as presented in Figure 2.
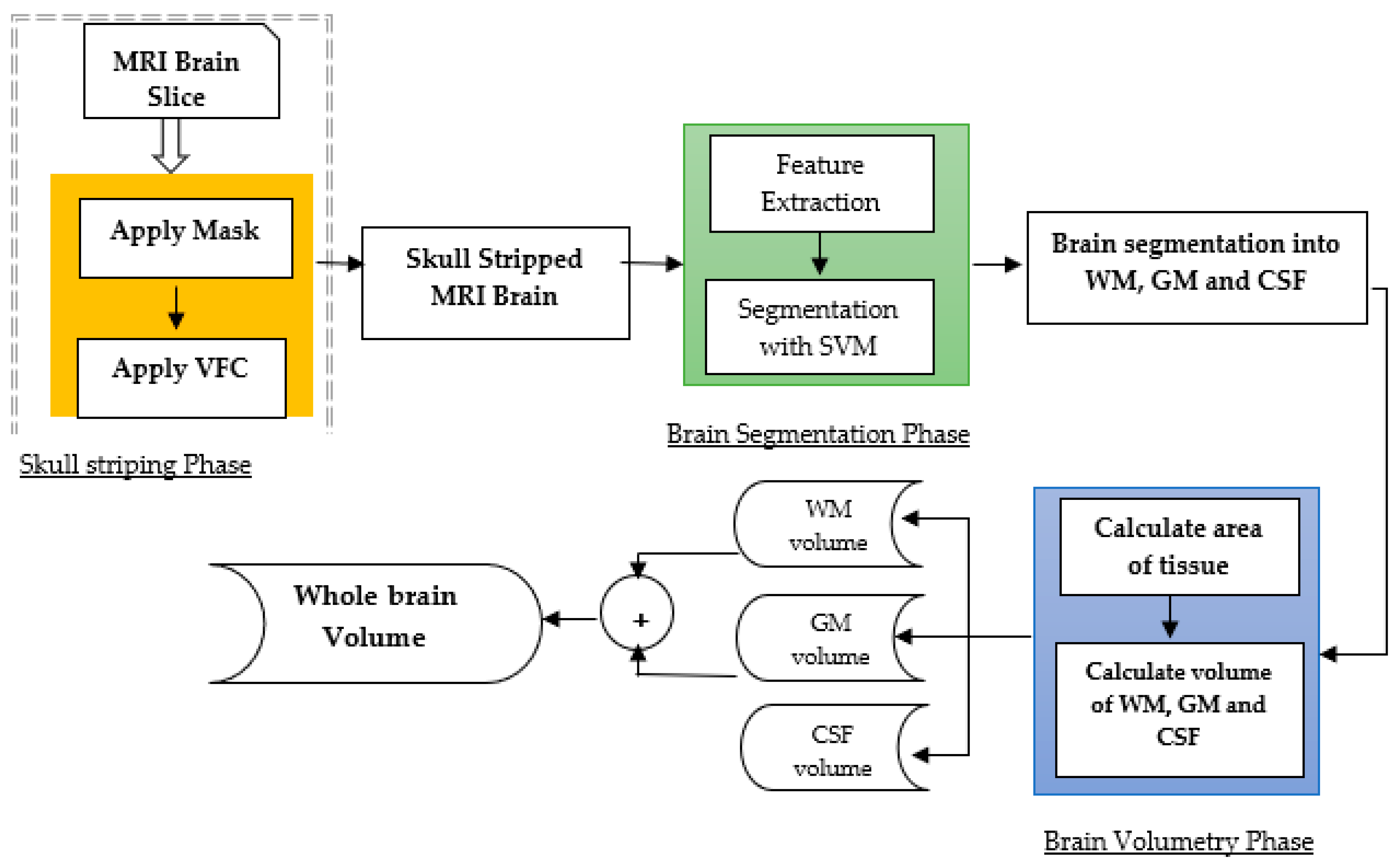
Figure 2. Model-based approach for brain tumor segmentation [26].
Several studies have conducted a comprehensive analysis of the model-based techniques used in brain tumor segmentation. Tandoori et al. [26] considered a couple of machine-learning-based models including random forests, decision trees, and support vector machines (SVMs), and reported that SVMs were the best models among all models. Similarly, Jiang et al. [27] presented a DNN model as a hybrid approach of the CNN and conditional random field and reported a very good performance.
Model-focused approaches can achieve high segmentation accuracy, but these have a substantial trade-off concerning the size of the dataset. For better performance, these models need a large high-quality dataset. However, a large dataset is also prone to data overfitting issues. Therefore, significant attention should be paid to the experimentation to obtain the best trade-off between the size of the dataset and the performance of model-focused techniques. In such cases, DL-based methods have revealed very good results in improving segmentation accuracy and robustness.
3. DL-Based Brain Tumor Segmentation
DL-based models, particularly convolutional neural networks, have demonstrated their effectiveness in medical imaging [28]. A traditional set up of a CNN-based DL model is presented in Figure 3. CNNs are designed to automatically learn the features that distinguish between tumor and non-tumor regions by processing the image using a series of kernels, convolutional and pooling layers, activations, and loss functions. CNNs have the advantage of being able to obtain features from the input, allowing them to capture complex patterns and variations in image intensity and texture [5].
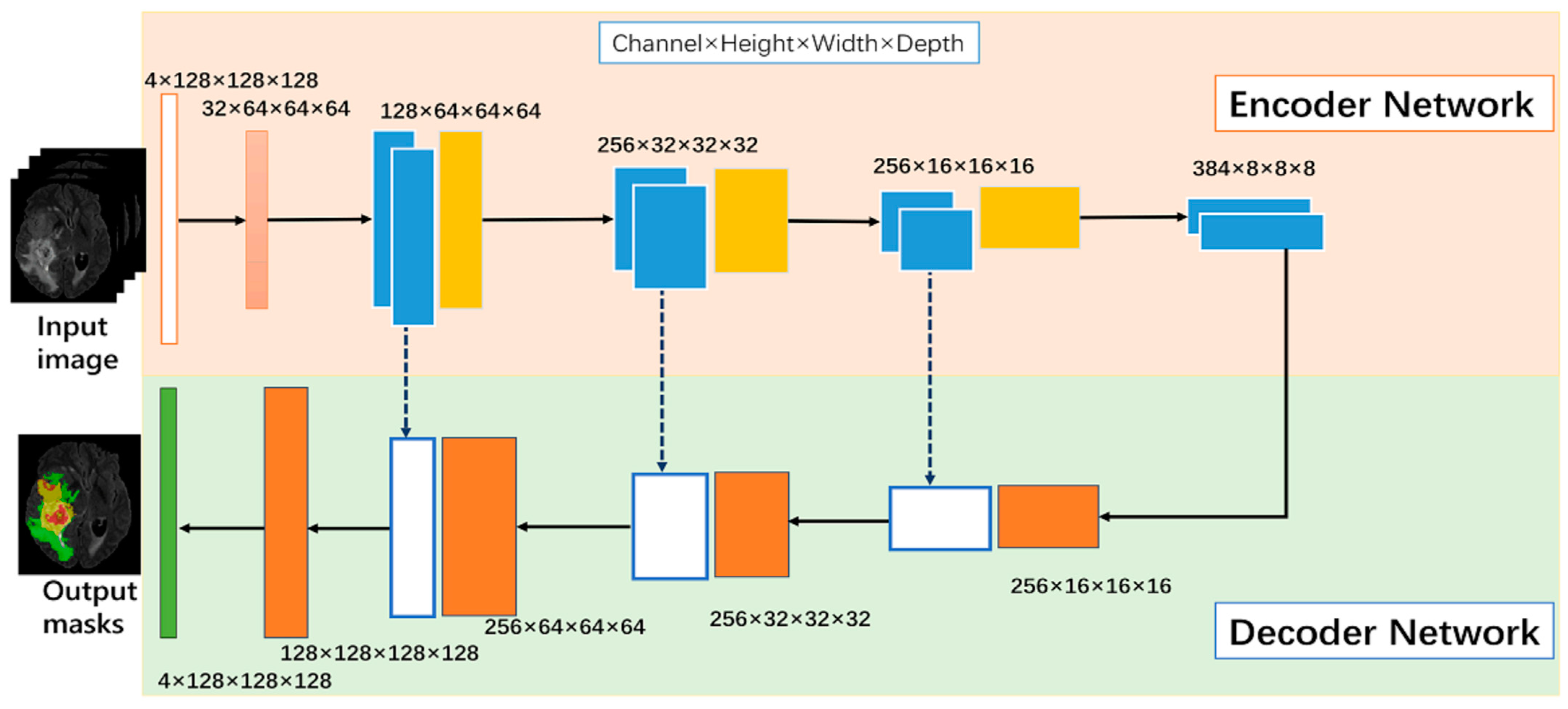
Figure 3. Neural-network-based model [28].
Several DL-based models have been developed for brain tumor segmentation, including V-Net, U-Net, and their variants. U-Net is a popular CNN architecture that consists of a contracting path to capture the contextual information of the input image and an expanding path to produce the segmentation map. V-Net is an extension of U-Net that includes a 3D CNN for processing volumetric data. These architectures have achieved state-of-the-art performance on various brain tumor segmentation challenges, such as the Multimodal Brain Tumor Segmentation Challenge (BRATS).
3.1. Neural-Network-Based Models
Several network structures, including CNNs and RNNs, have been proposed for brain tumor segmentation. CNNs are a type of deep neural network that has shown remarkable success in various image-processing tasks, including object recognition, segmentation, and classification. CNNs consist of multiple convolutional layers that extract the features of the input image and a fully connected layer that produces the segmentation map [29]. RNNs are another type of neural network that can process sequential data, such as time series or sequences of images. RNNs use feedback connections to retain information about the previous inputs and produce outputs that depend on the current and past inputs. RNNs have been applied to brain tumor segmentation by processing sequences of images and incorporating spatial and temporal information into the segmentation.
Several studies have compared the performance of different neural network structures for brain tumor segmentation. Akilan et al. [30] proposed a 3D CNN-LSTM and found that the LSTM architecture achieved the highest segmentation accuracy. Another study by Zhao et al. [31] proposed a CNN-RNN architecture that combines a 3D MRI brain tumor segmentation to improve segmentation accuracy.
CNNs and RNNs have been widely used for brain tumor segmentation due to their potential to learn intricate features from the input image and process sequential data. The choice of the most appropriate neural network structure depends on the specific application and available resources. Further research is needed to explore the potential of these structures in brain tumor segmentation.
3.2. Specialized Neural-Network-Based Model for Medical Imaging
U-Net and V-Net are two dedicated architectures that have been proposed for medical image segmentation, including brain tumor segmentation. U-Net is a fully convolutional neural network that involves an expanding path and a contracting path. The contracting path captures the context of the input image by reducing the spatial dimensions, while the expanding path produces the segmentation map by increasing the spatial dimensions [27].
Maqsood et al. [32] used fuzzy-logic-based edge detection and U-NET CNN classification. The approach involves preprocessing, edge detection, and feature extraction using wavelet transform, followed by CNN-based classification. The method outperforms other approaches in terms of accuracy and visual quality. Nawaz et al. [33] combine DenseNet77 in the encoder and U-NET in the decoder. The method achieves high segmentation accuracy on ISIC-2017 and ISIC-2018 datasets. Meraj et al. [34] used a quantization-assisted U-Net approach for accurate breast lesion segmentation in ultrasonic images. It combined U-Net segmentation with quantization and Independent Component Analysis (ICA) for feature extraction.
V-Net is an extension of U-Net that uses a 3D convolutional neural network for processing volumetric data. Isensee et al. [28] compared the performance of various deep-learning-based methods, including U-Net, V-Net, and other architectures, and found that U-Net achieved the best segmentation accuracy. The authors introduced a U-Net Cascade approach for large image datasets. Their approach contained two stages. Stage 1 trained a 3D U-Net on downsampled images; it enhanced the results of the image and fed it as inputs for Stage 2. Stage 2 used a second 3D U-Net for patch-based training at full resolution. Their two-stage structure is shown in Figure 4.
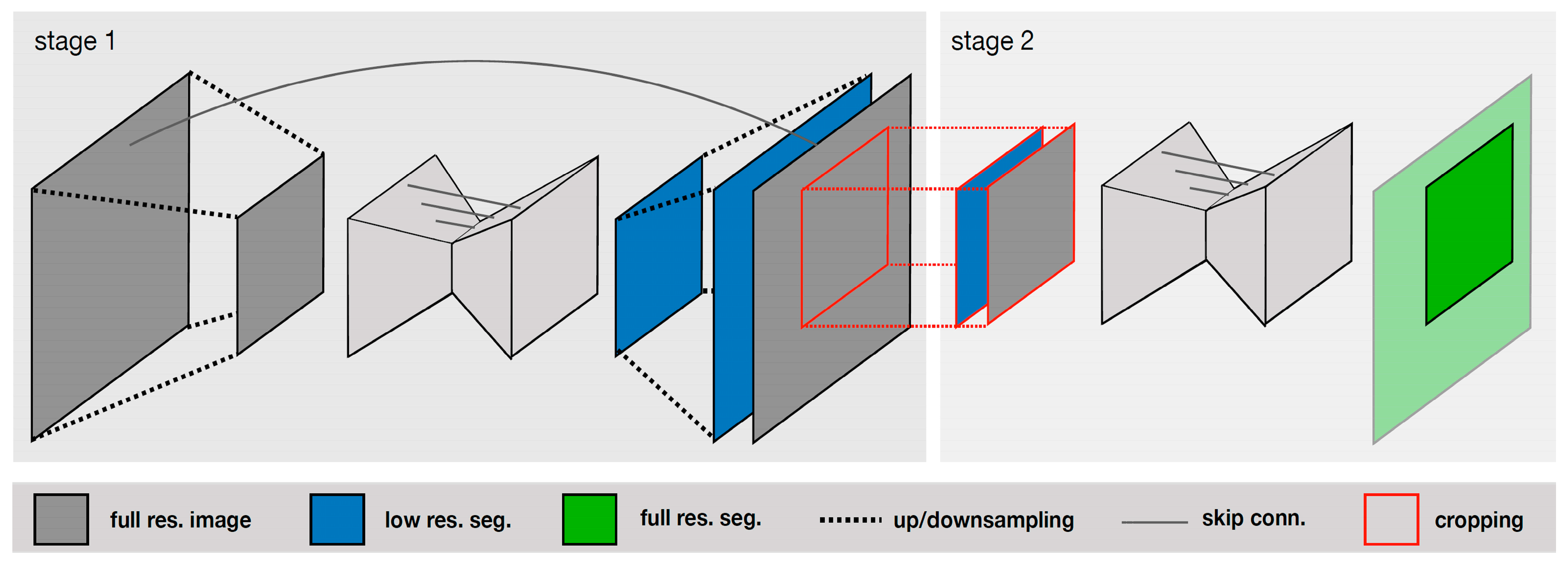
Figure 4. Specialized deep learning approaches [28].
Several studies have compared the performance of U-Net and V-Net with other deep-learning-based methods for brain tumor segmentation. Wang et al. [35] proposed a U-Net-based method that uses a multi-scale feature extraction strategy to improve segmentation accuracy. Other dedicated architectures, such as DeepMedic, BRATS-Net, and Tiramisu-Net, have also been proposed for brain tumor segmentation. DeepMedic is a hybrid CNN that combines a multi-scale architecture with a multi-path architecture to improve segmentation accuracy. BRATS-Net is a CNN architecture that includes a cascaded network and a refinement network for processing multimodal brain images. Tiramisu-Net is a densely connected convolutional network that uses skip connections to capture the features at different scales. Thus, there are several dedicated architectures that have also shown promising results in improving segmentation accuracy and robustness. The choice of the most appropriate architecture depends on the specific application and available resources.
4. Federated Learning for Medical Imaging
FL is a novel machine learning technique that enables multiple parties to collaboratively train a global model without sharing their sensitive data [36]. FL has been widely used in the context of medical imaging, where privacy and security concerns are of paramount importance. In FL, each participating client trains a local model on its data and shares only the model weights with the central server. The central server aggregates the model w and sends the updated global model back to the clients. FL allows medical institutions to collaborate and leverage their data to improve the accuracy and generalization of machine learning models while preserving the privacy and security of the data.
Several studies have demonstrated the effectiveness of FL in medical imaging tasks, including brain tumor segmentation. Khan et al. [37] proposed an FL-based framework for brain tumor segmentation that allowed multiple medical institutions to collaborate on the training of a deep learning model without sharing their data. The FL-based method achieved comparable segmentation accuracy to the centralized learning method while preserving the privacy and security of the data. Arikumar et al. [38] proposed an FL-based method for multimodal brain tumor segmentation that allowed multiple institutions to collaboratively train a global model using both MRI and histology data. The FL-based method achieved higher segmentation accuracy than the traditional centralized learning method while preserving the privacy and security of the data, as shown in Figure 5.
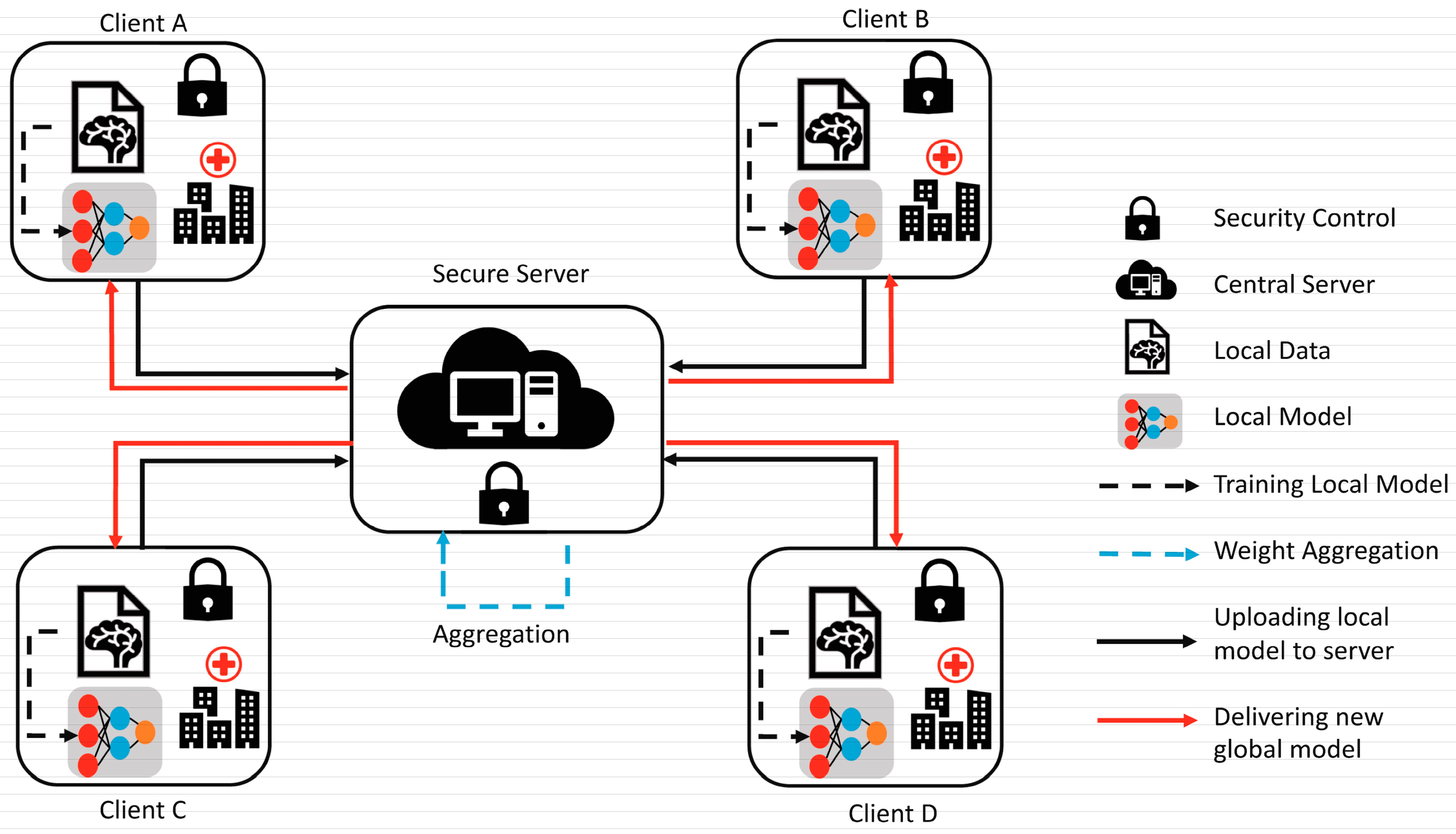
Figure 5. Federated learning [FL]-based model [37].
FL is a promising technique for medical tasks, including brain tumor segmentation, that enables multiple institutions to collaborate on the training of deep learning models without compromising the privacy and security of the data. Further research is needed to explore the potential of FL in medical imaging and to address the challenges related to the communication, computation, and heterogeneity of the data.
4.1. Data Confidentiality and Safety Concerns
Data confidentiality and safety concerns are critical issues in medical imaging, especially in the context of brain tumor segmentation, where patient data is highly sensitive and confidential. Deep-learning-based methods require large amounts of patient data for training, and the sharing of such data among multiple institutions raises concerns about data privacy, security, and ownership [39]. Several studies have addressed data confidentiality and safety concerns in medical imaging by proposing secure and privacy-preserving methods for data sharing and machine learning. Sheller et al. [40] proposed a secure data-sharing method that uses homomorphic encryption to enable multiple institutions to collaborate on the training of a machine learning model without revealing sensitive data. It achieved comparable performance to the traditional centralized learning method while preserving the privacy and security of the data. Wang et al. [35] proposed a blockchain-based method for sharing medical data among multiple institutions while maintaining data ownership and privacy. The authors demonstrated that the method provides a secure and transparent mechanism for data sharing and machine learning while preserving the privacy and security of the data.
Moreover, several studies have addressed the legal and ethical implications of data sharing and machine learning in medical imaging. Jiang et al. [41] proposed a framework for responsible data sharing in medical research that takes into account legal, ethical, and social considerations. Data confidentiality and safety concerns are critical issues in medical imaging, especially in the context of brain tumor segmentation. Secure and privacy-preserving methods, such as homomorphic encryption and blockchain, have been proposed to enable data sharing and machine learning while preserving the privacy and security of the data. Further research is needed to explore the potential of these methods in medical imaging and to address the legal and ethical implications of data sharing and machine learning.
4.2. Cooperative Learning for Better Performance
Cooperative learning, also known as collaborative learning, is a method that involves multiple learners working together to achieve a common goal. In the context of brain tumor segmentation, cooperative learning has been used to improve the performance of machine learning models by leveraging the collective intelligence of multiple experts [42]. Several studies have explored the potential of cooperative learning for brain tumor segmentation. Amiri et al. [43] proposed a cooperative learning framework that combines the outputs of multiple deep learning models to improve segmentation accuracy. The authors stated that the cooperative learning framework achieved higher segmentation accuracy than individual models and traditional ensemble learning methods. Witowski et al. [44] proposed a cooperative learning method that involves multiple experts in a crowdsourcing platform to annotate brain tumor images. The authors demonstrated that the cooperative learning method achieved higher annotation accuracy and consistency than individual experts and traditional methods.
Moreover, several studies have investigated the impact of cooperative learning on the interpretation and generalization of machine learning models. Abramoff et al. proposed a cooperative learning method that involves radiologists and deep learning models to jointly interpret brain MRI images. The proposed cooperative learning method achieved higher diagnostic accuracy and interpretability than individual radiologists and deep learning models.
Cooperative learning is a promising method for improving the performance and interpretability of machine learning models in brain tumor segmentation. Further research is needed to explore the potential of cooperative learning in other medical imaging tasks and to address the challenges related to the communication, computation, and heterogeneity of the data.
4.3. Federated Learning Applications
FL is a novel machine learning technique that has been applied in various domains, including healthcare, finance, and the internet of things (IoT). In healthcare, FL has been used for various tasks, such as disease diagnosis, drug discovery, and medical imaging analysis [45]. Chen et al. [46] proposed an FL-based framework for personalized cancer diagnosis that allowed multiple hospitals to collaboratively train a DL model without sharing their patient data. The FL-based method achieved higher accuracy than the traditional centralized learning method while preserving the privacy and security of the data. Dayan et al. [47] proposed a FL-based method for predicting COVID-19 severity using chest X-ray images. The authors showed that the FL-based method achieved higher accuracy than the traditional centralized learning method while preserving the privacy and security of the data.
Moreover, several studies have explored the potential of FL for medical image analysis tasks, such as brain tumor segmentation, retinal image analysis, and skin lesion classification. Zhang et al. [48] present an FL-based method for retinal image analysis that allowed multiple medical institutions to collaborate on the training of a deep learning model without sharing their patient data. The authors demonstrated that the FL-based method achieved higher accuracy than the traditional centralized learning method while preserving data security privacy.
FL is a promising technique for various ML applications, including healthcare and medical imaging analysis. However, further research is needed to explore the potential of FL in other domains and to address the challenges related to the communication, computation, and heterogeneity of the data.
This entry is adapted from the peer-reviewed paper 10.3390/math11194189
References
- Louis, D.N.; Perry, A.; Reifenberger, G.; Von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820.
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans. Med. Imaging 2014, 34, 1993–2024.
- Huguet, E.E.; Huyhn, E.; Berry, C.R. Computed Tomography and Magnetic Resonance Imaging. Atlas Small Anim. Diagn. Imaging 2023, 14, 16–26.
- Bakas, S.; Reyes, M.; Jakab, A.; Bauer, S.; Rempfler, M.; Crimi, A.; Shinohara, R.T.; Berger, C.; Ha, S.M.; Rozycki, M.; et al. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. arXiv 2018, arXiv:1811.02629.
- Liu, Z.; Tong, L.; Chen, L.; Jiang, Z.; Zhou, F.; Zhang, Q.; Zhang, X.; Jin, Y.; Zhou, H. Deep learning based brain tumor segmentation: A survey. Complex Intell. Syst. 2023, 9, 1001–1026.
- Ullah, F.; Salam, A.; Abrar, M.; Amin, F. Brain Tumor Segmentation Using a Patch-Based Convolutional Neural Network: A Big Data Analysis Approach. Mathematics 2023, 11, 1635.
- Shankar, R.S.; Raminaidu, C.; Raju, V.S.; Rajanikanth, J. Detection of Epilepsy based on EEG Signals using PCA with ANN Model. J. Phys. Conf. Ser. 2021, 1, 012145.
- Singh, S.; Mittal, N.; Singh, H. A feature level image fusion for IR and visible image using mNMRA based segmentation. Neural Comput. Appl. 2022, 34, 8137–8154.
- Wu, J.; Zhou, L.; Gou, F.; Tan, Y. A Residual Fusion Network for Osteosarcoma MRI Image Segmentation in Developing Countries. Comput. Intell. Neurosci. 2022, 2022, 7285600.
- Wang, A.; Zhang, Q.; Han, Y.; Megason, S.; Hormoz, S.; Mosaliganti, K.R.; Lam, J.C.K.; Li, V.O.K. A novel deep learning-based 3D cell segmentation framework for future image-based disease detection. Sci. Rep. 2022, 12, 342.
- Hamza, R.; Hassan, A.; Ali, A.; Bashir, M.B.; Alqhtani, S.M.; Tawfeeg, T.M.; Yousif, A. Towards Secure Big Data Analysis via Fully Homomorphic Encryption Algorithms. Entropy 2022, 24, 519.
- Li, D.; Han, D.; Weng, T.-H.; Zheng, Z.; Li, H.; Liu, H.; Castiglione, A.; Li, K.-C. Blockchain for federated learning toward secure distributed machine learning systems: A systemic survey. Soft Comput. 2022, 26, 4423–4440.
- Dang, T.K.; Lan, X.; Weng, J.; Feng, M. Federated learning for electronic health records. ACM Trans. Intell. Syst. Technol. (TIST) 2022, 13, 1–17.
- Brisimi, T.S.; Chen, R.; Mela, T.; Olshevsky, A.; Paschalidis, I.C.; Shi, W. Federated learning of predictive models from federated Electronic Health Records. Int. J. Med. Inform. 2018, 112, 59–67.
- Xu, J.; Glicksberg, B.S.; Su, C.; Walker, P.; Bian, J.; Wang, F. Federated learning for healthcare informatics. J. Healthc. Inform. Res. 2021, 5, 1–19.
- Gordillo, N.; Montseny, E.; Sobrevilla, P. State of the art survey on MRI brain tumor segmentation. Magn. Reson. Imaging 2013, 31, 1426–1438.
- A Okuboyejo, D.; O Olugbara, O. A Review of Prevalent Methods for Automatic Skin Lesion Diagnosis. Open Dermatol. J. 2018, 12, 14–53.
- Kimberlin, C.L.; Winterstein, A.G. Validity and reliability of measurement instruments used in research. Am. J. Health-Syst. Pharm. 2008, 65, 2276–2284.
- Kavitha, A.; Chellamuthu, C.; Rupa, K. An efficient approach for brain tumour detection based on modified region growing and neural network in MRI images. In Proceedings of the 2012 international conference on Computing, Electronics and Electrical Technologies (ICCEET), Nagercoil, India, 21–22 March 2012; pp. 1087–1095.
- Singh, J.F.; Magudeeswaran, V. Thresholding based method for segmentation of MRI brain images. In Proceedings of the 2017 International Conference on I-SMAC (IoT in Social, Mobile, Analytics and Cloud) (I-SMAC), Palladam, India, 10–11 February 2017; pp. 280–283.
- Jaglan, P.; Dass, R.; Duhan, M. A comparative analysis of various image segmentation techniques. In Proceedings of the 2nd International Conference on Communication, Computing and Networking: ICCCN 2018, NITTTR, Chandigarh, India, 29–30 March 2018; pp. 359–374.
- Charutha, S.; Jayashree, M.J. An efficient brain tumor detection by integrating modified texture based region growing and cellular automata edge detection. In Proceedings of the 2014 International conference on control, instrumentation, communication and computational technologies (ICCICCT), Kanyakumari District, India, 10–11 July 2014; pp. 1193–1199.
- Rajan, P.G.; Sundar, C. Brain Tumor Detection and Segmentation by Intensity Adjustment. J. Med. Syst. 2019, 43, 1–13.
- Sheela, C.J.J.; Suganthi, G. Morphological edge detection and brain tumor segmentation in Magnetic Resonance (MR) images based on region growing and performance evaluation of modified Fuzzy C-Means (FCM) algorithm. Multimedia Tools Appl. 2020, 79, 17483–17496.
- Unde, A.S.; Premprakash, V.A.; Sankaran, P. A novel edge detection approach on active contour for tumor segmentation. In Proceedings of the 2012 Students Conference on Engineering and Systems, Allahabad, India, 16–18 March 2012; pp. 1–6.
- Tanoori, B.; Azimifar, Z.; Shakibafar, A.; Katebi, S. Brain volumetry: An active contour model-based segmentation followed by SVM-based classification. Comput. Biol. Med. 2011, 41, 619–632.
- Jiang, J.; Zhang, Z.; Huang, Y.; Zheng, L. Incorporating depth into both cnn and crf for indoor semantic segmentation. In Proceedings of the 2017 8th IEEE International Conference on Software Engineering and Service Science (ICSESS), Beijing, China, 24–26 November 2017; pp. 525–530.
- Someswararao, C.; Shankar, R.S.; Appaji, S.V.; Gupta, V. Brain tumor detection model from MR images using convolutional neural network. In Proceedings of the 2020 International Conference on System, Computation, Automation and Networking (ICSCAN), Pondicherry, India, 3–4 July 2020; pp. 1–4.
- Zhao, L.; Jia, K. Multiscale CNNs for Brain Tumor Segmentation and Diagnosis. Comput. Math. Methods Med. 2016, 2016, 8356294.
- Akilan, T.; Wu, Q.J.; Safaei, A.; Huo, J.; Yang, Y. A 3D CNN-LSTM-Based Image-to-Image Foreground Segmentation. IEEE Trans. Intell. Transp. Syst. 2019, 21, 959–971.
- Zhao, Y.; Ren, X.; Hou, K.; Li, W. Recurrent Multi-Fiber Network for 3D MRI Brain Tumor Segmentation. Symmetry 2021, 13, 320.
- Maqsood, S.; Damasevicius, R.; Shah, F.M. An Efficient Approach for the Detection of Brain Tumor Using Fuzzy Logic and U-NET CNN Classification; Springer International Publishing: Cham, Switzerland, 2021; pp. 105–118.
- Nawaz, M.; Nazir, T.; Masood, M.; Ali, F.; Khan, M.A.; Tariq, U.; Sahar, N.; Damaševičius, R. Melanoma segmentation: A framework of improved DenseNet77 and UNET convolutional neural network. Int. J. Imaging Syst. Technol. 2021, 32, 2137–2153.
- Meraj, T.; Alosaimi, W.; Alouffi, B.; Rauf, H.T.; Kumar, S.A.; Damaševičius, R.; Alyami, H. A quantization assisted U-Net study with ICA and deep features fusion for breast cancer identification using ultrasonic data. PeerJ Comput. Sci. 2021, 7, e805.
- Wang, G.; Li, W.; Ourselin, S.; Vercauteren, T. Automatic Brain Tumor Segmentation Based on Cascaded Convolutional Neural Networks With Uncertainty Estimation. Front. Comput. Neurosci. 2019, 13, 56.
- Malik, H.; Anees, T.; Naeem, A.; Naqvi, R.A.; Loh, W.-K. Blockchain-Federated and Deep-Learning-Based Ensembling of Capsule Network with Incremental Extreme Learning Machines for Classification of COVID-19 Using CT Scans. Bioengineering 2023, 10, 203.
- Khan, M.I.; Jafaritadi, M.; Alhoniemi, E.; Kontio, E.; Khan, S.A. Adaptive Weight Aggregation in Federated Learning for Brain Tumor Segmentation. In Proceedings of the Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: BrainLes 2021, Held in Conjunction with MICCAI 2021, Virtual Event, 27 September 2021; Revised Selected Papers, Part II. pp. 455–469.
- Arikumar, K.S.; Prathiba, S.B.; Alazab, M.; Gadekallu, T.R.; Pandya, S.; Khan, J.M.; Moorthy, R.S. FL-PMI: Federated Learning-Based Person Movement Identification through Wearable Devices in Smart Healthcare Systems. Sensors 2022, 22, 1377.
- Shini, S.; Thomas, T.; Chithraranjan, K. Cloud Based Medical Image Exchange-Security Challenges. Procedia Eng. 2012, 38, 3454–3461.
- Sheller, M.J.; Edwards, B.; Reina, G.A.; Martin, J.; Pati, S.; Kotrotsou, A.; Milchenko, M.; Xu, W.; Marcus, D.; Colen, R.R.; et al. Federated learning in medicine: Facilitating multi-institutional collaborations without sharing patient data. Sci. Rep. 2020, 10, 12598.
- Jiang, X.; Zhou, X.; Grossklags, J. Privacy-Preserving High-dimensional Data Collection with Federated Generative Autoencoder. Proc. Priv. Enhancing Technol. 2022, 2022, 481–500.
- Luo, G.; Zhang, H.; He, H.; Li, J.; Wang, F.-Y. Multiagent Adversarial Collaborative Learning via Mean-Field Theory. IEEE Trans. Cybern. 2020, 51, 4994–5007.
- Amiri, S.; Mahjoub, M.A.; Rekik, I. Bayesian Network and Structured Random Forest Cooperative Deep Learning for Automatic Multi-label Brain Tumor Segmentation. ICAART 2018, 2, 183–190.
- Witowski, J.; Choi, J.; Jeon, S.; Kim, D.; Chung, J.; Conklin, J.; Longo, M.G.F.; Succi, M.D.; Do, S. MarkIt: A Collaborative Artificial Intelligence Annotation Platform Leveraging Blockchain For Medical Imaging Research. Blockchain Health Today 2021, 4, 11–15.
- Joshi, M.; Pal, A.; Sankarasubbu, M. Federated learning for healthcare domain-Pipeline, applications and challenges. ACM Trans. Comput. Healthc. 2022, 3, 1–36.
- Ma, Q.; Zhou, S.; Li, C.; Liu, F.; Liu, Y.; Hou, M.; Zhang, Y. DGRUnit: Dual graph reasoning unit for brain tumor segmentation. Comput. Biol. Med. 2022, 149, 106079.
- Dayan, I.; Roth, H.R.; Zhong, A.; Harouni, A.; Gentili, A.; Abidin, A.Z.; Liu, A.; Costa, A.B.; Wood, B.J.; Tsai, C.-S.; et al. Federated learning for predicting clinical outcomes in patients with COVID-19. Nat. Med. 2021, 27, 1735–1743.
- Zhang, M.; Qu, L.; Singh, P.; Kalpathy-Cramer, J.; Rubin, D.L. SplitAVG: A Heterogeneity-Aware Federated Deep Learning Method for Medical Imaging. IEEE J. Biomed. Health Inform. 2022, 26, 4635–4644.
This entry is offline, you can click here to edit this entry!
