1. Virus-Induced Gene Silencing (VIGS) System
Vegetables are grown worldwide and play an important role in the nutrition demands of humans’ daily diets, especially for providing vitamins, minerals, and dietary fiber, which have been strongly associated with human health. They can also be a major source of protein in poor regions. The continuous increase in human living standards, along with increasing demand for vegetable production and quality, make the improvement of molecular breeding technologies applied to vegetable breeding a need to achieve more efficient and sustainable crop production. This market demand for high-quality and more uniform products, together with global warming, oblige scientists to explore gene functions which are important for agronomic traits (e.g., disease, pest, or abiotic stress resistance) in vegetables. Traditional methods to study plant gene function include transgenic technology, gene knockout, gene-induced overexpression, and RNAi technology. These research methods have certain limitations, such as long research cycles, the need for genetic transformation, and low conversion efficiency, limiting rapid and efficient study of plant gene functions
[1][2]. However, Virus-Induced Gene Silencing (VIGS) provides an alternative tool to investigate gene functional validation in vegetables.
VIGS is an effective method for switching off the expression of a gene. It was developed based on the mechanism of plants’ defenses against viruses, using RNA-mediated post-transcriptional gene silencing (PTGS)
[3][4][5]. It has emerged rapidly as a key regulator of gene expression applicable to reverse genetics for plant gene functional studies. Plant scientists discovered gene silencing-related mechanisms while performing plant transformation experiments in which the introduction of a transgene into a desired genome resulted in the silencing of both the transgene and its homologous endogenes
[6][7]. As a result of these first observations, plant geneticists and biologists developed this molecular biology approach to address not only gene silencing method questions, but also to explore the complexity of the biological pathways involved, as well as to demonstrate their multilayer relationships with one another. For instance, it is well documented that, after the virus infests a plant, viral transcription and replication in the plant cell cytoplasm lead to a double-stranded RNA (dsRNA), which is key in the VIGS process. dsRNA was cleaved into small interfering RNA (siRNA) by the Dicer or Dicer-like (DCL) nuclease, ranging from 21 to 24 integrated nucleotides, and siRNA binds to RNase in plants as a single strand to form an RNA-induced silencing complex (RISC). Further, the RNA-induced silencing complexes (RISCs) cleave to viral RNA in the cytoplasm in a nucleotide-specific manner, ultimately triggering the degradation of the targeted mRNA
[8][9][10][11][12] (
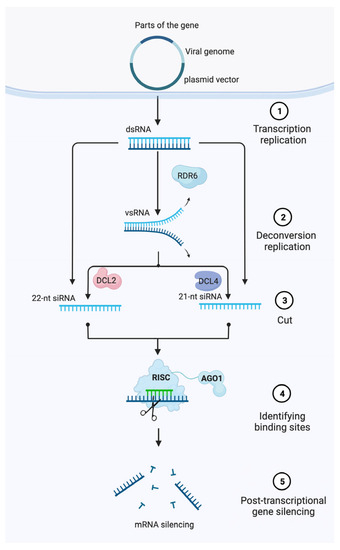
Figure 1). RNA silencing is an evolutionarily conserved RNA-mediated process
[13], where sequence-specific eukaryotic gene silencing mechanisms are involved in numerous biological processes in plants and animals
[14]. Therefore, virus infection has been proven as an efficient trigger of RNA silencing, turning VIGS into a powerful tool for gene function studies and vegetable improvement.
Figure 1. Model of post-transcriptional gene silencing (PTGS)-mediated gene silencing in plants. First, a partial segment (~200–500 bp) homologous to the target gene of interest is cloned into a modified single/multipartite viral genome harbored within a plasmid vector. Then, Agrobacteria are used to transfect plant cells and transfer DNA from the binary vector into the nucleus where it is transiently expressed. Finally, dsRNA formed during virus replication are cleaved by DICER proteins to produce siRNAs that guide RISC and the local amplification of siRNAs that systemically spread to induce post-transcriptional gene silencing of a target mRNA. Abbreviations: RISC, RNA-induced silencing complex; siRNA, small-interfering RNA; dsRNA, double-stranded RNA; DCL, DICER-Like.
2. Advantages of VIGS
2.1. Transient Silence of VIGS
Transient silencing of VIGS to analyze plant endogenous gene function is a fast and effective reverse genetic tool in plant functional genomics. This is possible since phenotypic changes induced by the down-regulation of endogenous plant genes can be detected in a short period of time
[1][15][16]. The effective silencing time and the effectiveness of viral vectors differ depending on the viral vector and the target plant infested
[2][17][18]. For instance, photobleaching in leaves, stems, axillary buds, and sepals of Tobacco Rattle Virus (TRV)-based VIGS system, using
phytoene desaturase (
PDS) as a reporter gene, infiltrated tobacco (
Nicotiana benthamiana) plants appeared 10 days after TRV infestation
[19]. Another observation was that the percentage of white tissue in leaves decreased 28 days after infestation
[5]. A similar experiment in tomato reported that the transient silencing response of TRV–
PDS sprayed on 4-week-old tomato seedlings showed symptoms of photobleaching caused by
Silene latifolia (
Sl)
PDS gene silencing after ~8 weeks of leaf inoculation. The systemic photobleaching persisted throughout the experiment for 4 months after the inoculation
[19].
In another experiment to assess silencing by
Agrobacterium-mediated barley stripe mosaic virus (BSMV) VIGS, a 370 bp PDS (
NbPDS) fragment from
N.benthamiana was cloned into pCa-cbLIC to generate pCa-cb:NbPDS370. Then, the four-leaf stage of
N. benthamiana was infiltrated with Agrobacterium mixtures containing pCaBS-a, pCaBS-b, and pCa-cb: NbPDS370. Leaves infiltrated with virus to elicit PDS silencing developed a mottled photobleaching phenotype on the fifth or sixth leaves at 9 to 10 dpi, and, about 5 days later (15 dpi), larger (and more uniform) white PDS silencing areas were observed at the 6- to 8-leaf-stage. Furthermore, PDS silencing was most pronounced at 30 to 45 dpi, with larger and more apparent areas of photobleaching on many stems and petioles
[20].
2.2. VIGS Overcomes Functional Redundancy
Determination of gene function is particularly problematic when studying large gene families because two or more genes could perform the same function, either by gene copy duplication or a higher ploidy level. The inactivation of one of these genes has little or no effect on the phenotypic appearance; thus, gene redundancy limits the ability to experimentally assess the contributions of individual genes. However, VIGS can overcome this gene function redundancy by constructing the viral vector carrying highly conserved regions of the target gene family and potentially knocking off all the family members
[1][21]. One example is the heat shock protein 90 (
HSP90) that belongs to a large gene family of transcription factors that control fundamental processes of plant development. An insertion of the highly conserved coding sequence of the
HSP90 gene family into the Potato Virus X (PVX) viral vector silenced all
HSP90 mRNAs and was confirmed by protein blotting in tomato. Lack of HSP90 protein led to stunted development and leaf deformation plant phenotypes.
VIGS can also overcome the redundancy issue in polyploid species. Cabbage (
Brassica rapa L.) is a globally significant vegetable crop (71 million tonnes per year)
[22], where its ploidy level, high gene duplication rate, and long growth cycle have posed challenges for stable genetic transformation, greatly limiting study at the gene functional level. To overcome this challenge, a VIGS system of cabbage was constructed using the Tomato Yellow Leaf Curl Virus (TYLCV) viral vector.
VIGS was also used to investigate the role of transcription factors (TFs) synchronized with the expression of genes related to programmed cell death (PCD) during PCD and salt stress. Knockdown mutants of these TFs were generated in tobacco by modifying the TRV and utilizing VIGS to produce knockout mutants of these TFs in tobacco. Results of knockdown mutant tobacco cells confirmed the influence of two TFs during PCD. In addition, the knockout insertion mutants and overexpression lines indicated the role of ERF109 in conferring salt tolerance in
Arabidopsis [23].
2.3. VIGS Overcomes Conditional Constraints
CRISPR/Cas9 technology is widely used for gene validation by performing gene knockouts at the DNA level. Although it is a powerful technology, it may not be suitable when investigating essential genes that have been shown to be plantlet lethal (in the knockout stage) during the regeneration of plant transformation
[21]. The main advantage of VIGS is that it can effectively down-regulate the expression of those same essential genes and can provide a better understanding of gene effects’ influence on the phenotype, primarily by taking advantage of post-translation regulation impacts of reducing protein level expression
[1][2][24]. Another benefit is that, because the knockdown regulation is temporary, it can return to normal growth and seed production and does not retain the virus or vector components
[25].
One example that highlights the power of VIGS as a tool to study temporarily inhibited gene expression is the
Proliferating Cell Nuclear Antigen (
PCNA), which is essential for host cell growth and development. For
PCNA, most mutations are lethal and difficult to retrieve. Therefore, gene functional verification cannot be performed by transgenic silencing.
PCNA is an important component in the replication and repair machinery involved in nucleic acid metabolism
[26]. PCNA contributes to the persistent DNA polymerase δ and DNA polymerase ε synthesis factor that attaches the polymerase catalytic unit to the DNA template for rapid and sustained DNA synthesis. Knockout of
PCNA in plants by CRISPR/Cas9 methods leads to death during regeneration, providing only partial information on the gene function due to the scarcity of phenotypes
[27]. In contrast, using VIGS technology to silence the
PCNA gene in tomato permitted the screening of the whole set of individuals tested. This essay resulted in severely stunted growth of infested tomato plants with the VIGS–PCNA viral vector, in contrast to no morphological effects observed in an empty vector plant test with VIGS–GFP as the reported gene. This proved the importance of the virus-induced gene silencing technology in demonstrating the causality of the
PCNA gene in tomatoes
[28].
2.4. Disadvantages VIGS
When performing a comparison of VIGS technologies, their main disadvantages are that most viruses used for VIGS have a limited number of hosts, and the virus–host combination seems to be a crucial factor in determining the efficacy of silencing. Some of the viruses used in VIGS can cause symptoms that might mask the phenotype caused by the silencing of the target gene. Moreover, many viruses do not infect the growing points or floral parts of plants, especially the seed, precluding gene silencing in these tissues
[29].
3. VIGS Applications in Vegetable Plants
To date, many plant viruses have been successfully modified as VIGS vectors to induce targeted gene silencing in host vegetable plants, such as tobacco mosaic virus (TMV), PVX, and TRV. Among them, TRV is especially widely used in Solanaceae vegetables, and gene silencing can be effectively induced by constructing recombinant TRV virus vectors
[10][30][31].
TRV vector has been successfully applied in several plant organs (leaf, root, and flower), affecting key aspects of plant nutritional growth and reproductive stages
[5]. Recently, studies have shown that this same technology can be applied to fruits, for example, tomato or pepper
[32]. In tomato, the characteristic bleaching phenotype after TRV–PDS injection was obtained and those symptoms expanded, infesting peduncles at the tomato fruit developmental stage. Gene silencing was confirmed at the molecular level by qPCR. In pepper, an optimized TRV vector was developed using a Viral Silencing suppressor of RNA silencing (VSR). pTRV2-C2b-CaCCS vector was constructed, targeting a key gene in capsanthin/capsorubin biosynthesis that achieved high efficiency of calcium-activated chloride channels’ (CaCCS) protein silencing
[32]. Another example of studying fruit organs was the silencing of the tomato ethylene (EIN3)-binding F-box genes.
SlEBF1 and
SlEBF2 have been reported to negatively regulate ethylene signaling, causing constitutive ethylene-related symptoms, fertility defects, growth decline, plant senescence acceleration, and fruit ripening
[33]. Altogether, these examples show the impact this molecular gene silencing advancement can have, to better explain gene function validation throughout the whole vegetable life cycle.
VIGS applications in vegetables have been challenged by the host-range reduced diversity. As a matter of fact, TRV’s host-range reduced diversity has restricted the ability to test gene silencing effects in the Cucurbitaceae family. The discovery and modification ability of plant viruses has allowed using a broader host range of target vegetables, e.g., apple latent spherical virus (ALSV), tobacco ringspot virus (TRSV), cucumber green mottle mosaic virus (CGMMV), and tomato leaf curl virus (ToLCV). ALSV has a wide range of vegetable hosts, including the Solanaceae, the Cucurbitaceae, and the Fabaceae families, most of which have shown no viral symptoms. At the same time, this viral vector was shown to effectively induce stable virus-induced gene silencing in a wide range of vegetable plants and it has been shown to possess long-lasting effects. For example, in pea (
Pisum sativum L.), a 300 bp fragment of a PDS gene from soybean plants was inserted into ALSV-RNA2 vectors, and the resulting viruses (soyPDS-ALSV) were inoculated into primary leaves of pea plants. Inoculated pea plants initiated the development of white spots on the third trifoliate true leaf at 10 to 14 dpi and then showed highly uniform white photobleached phenotype in the fourth or fifth true leaves, indicating the PDS gene was silenced. The PDS silencing on these plants persisted for a month
[34]. This caused pea death after one month due to the lack of photosynthesis ability. Similar results were obtained when ALSV-CuPDS and ALSV-CuSU vectors were used to infect Cucurbitaceae plants, including pumpkin (
Cucurbita maxima L.) having mRNA 76% lower expression levels in the leaf tissues compared to controls after infection
[35]. However, one of the disadvantages of the ALSV vector comes from its gene expression strategy of the virus genome. As the proteins encoded by the ALSV genome are expressed by polyprotein synthesis followed by proteolytic processing, it is necessary to ligate target sequences in the frame to the cloning sites of the ALSV vector. This necessity makes it difficult to apply an ALSV vector for high throughput functional genomics, as reported by other vectors
[1][2][36].
Another viral vector with a wide host in vegetables is the tobacco ringspot virus (TRSV), a single-stranded positive-sense polyadenylated RNA molecules. This viral vector was first applied to cucurbits and legumes, having silenced all plants with new white leaves, petioles, and even tendrils being almost completely white
[37]. Furthermore, the silencing phenotype of the
PDS gene was stable and persisted for approximately 1 month
[35]. Recent studies have shown the role of the soybean mid–late flow protein gene (
GmLATE) in soybean by infesting plants with the VIGS system of TRSV. Researchers concluded that the silencing of
GmLATE reduced the expression of flowering-related genes and the arrest of flower development in soybean
[38]. They also demonstrated that the silencing effect of this virus vector can remain effective until the reproductive growth stages.
Unlike double-stranded RNA viruses, CGMMV is a positive-sense single-stranded RNA virus with a limited host range that turned out to be able to infect cucurbits
[12]. In recent reports, the photobleaching caused by the infection of CGMMV-PDS vector was observed on the third leaf of melon and gourd, and the fifth true leaf of cucumbers. The stability of the photobleaching was variable in watermelon, melon, and cucumber plants at 32, 20, and 39 days, respectively. However, the remaining challenge is that the silencing effect is not as evident in the whole tissue as shown in Liu et al.
[12], where the photobleaching phenotype was constrained at the vicinity of leaf veins. The positive side of this technique is the relative easiness of genetic manipulation of the virus vector, making this technique widely utilized in functional genomics on the Cucurbitaceae crop family.
ToLCV and PVX are two additional virus vectors with a wide host range that can be used to verify the gene function in tomatoes. The ToLCV vector belongs to the genus begomoviridae of the family Geminiviridae and was used to silence the PCNA endogenous gene in tomato, resulting in substantial stunting of the plant growth. Interestingly, the vector’s silencing effectiveness was enhanced with the inclusion of a mutation in the silencing suppressor Open Reading Frame (ORF) AC2
[28]. As for PVX, it has been used as a VIGS virus in the Solanaceous genus for a long time
[39]. For instance, it was used to study the role SlymiR157 has during the ripening process in tomato. Pre-SlymiR157 was cloned into a PVX-based VIGS vector to produce a PVX/pre-SlymiR157, obtaining a PVX able to efficiently deliver pre-SlymiR157 into fruits
[40].
This entry is adapted from the peer-reviewed paper 10.3390/horticulturae9080934