Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
MicroRNA (miRNA) are small noncoding RNAs that play vital roles in post-transcriptional gene regulation by inhibiting mRNA translation or promoting mRNA degradation. The dysregulation of miRNA has been implicated in numerous human diseases, including cancers. miR-34 family members (miR-34s), including miR-34a, miR-34b, and miR-34c, have emerged as the most extensively studied tumor-suppressive miRNAs.
- microRNA
- miR-34
- tumor suppressor
- miRNA therapy
1. Biosynthesis of MicroRNA-34 Family Members and Regulation of miR-34 Expression
1.1. Biosynthesis of MicroRNA-34 Family Members
The miR-34 family comprises three members, miR-34a, miR-34b, and miR-34c, encoded by genes located on chromosomes 1 and 11 [1]. MiR-34b and miR-34c share a common primary seed sequence on chromosome 11, while miR-34a is encoded on chromosome 1. Comparative analysis shows that miR-34a shares 86% identity with miR-34b and 82% with miR-34c [1][2][3]. miR-34a and miR-34c also share identical seed sequences with the miR-449 superfamily, including miR-449a, miR-449b, and miR-449c. miR-34a typically exhibits higher expression levels than miR-34b and miR-34c in most human cells [4].
The biosynthesis of miR-34 family members (miR-34s) is a multistep process that occurs in the nucleus and cytoplasm. RNA polymerase II or III transcribes miR-34 genes in the nucleus, generating a long hairpin-shaped molecule known as pri-miRNA. The DROSHA endonuclease cleaves the pri-miRNA into the 80–100-nucleotide-long pre-miRNAs within the nucleus. Exportin-5 transports the pre-miR-34s to the cytoplasm, where the DICER endonuclease further processes them into double-stranded mature miR-34s, approximately 20–23 nucleotides long [5]. These mature miRNAs associate with Argonaute proteins to form the RNA-induced silencing complex (RISC). One strand becomes the mature miRNA, while the other is degraded. miRNA-mediated gene silencing depends on the level of complementarity between the miR-34 seed sequence and target mRNA binding sites. Full complementarity may lead to mRNA degradation, while partial complementarity inhibits its translation [2][6][7].
The alignment of mature miR-34a, miR-34b, and miR-34c sequences is depicted in Figure 1. It is worth mentioning that the differential expression levels of miR-34s indicate their distinct and specific functions in the regulation of gene expression. The precise mechanisms through which miRNAs exert their effects on gene expression are still under investigation, and ongoing research endeavors are expanding our knowledge of the intricate and dynamic roles played by miRNAs in cellular physiology and development of diseases.
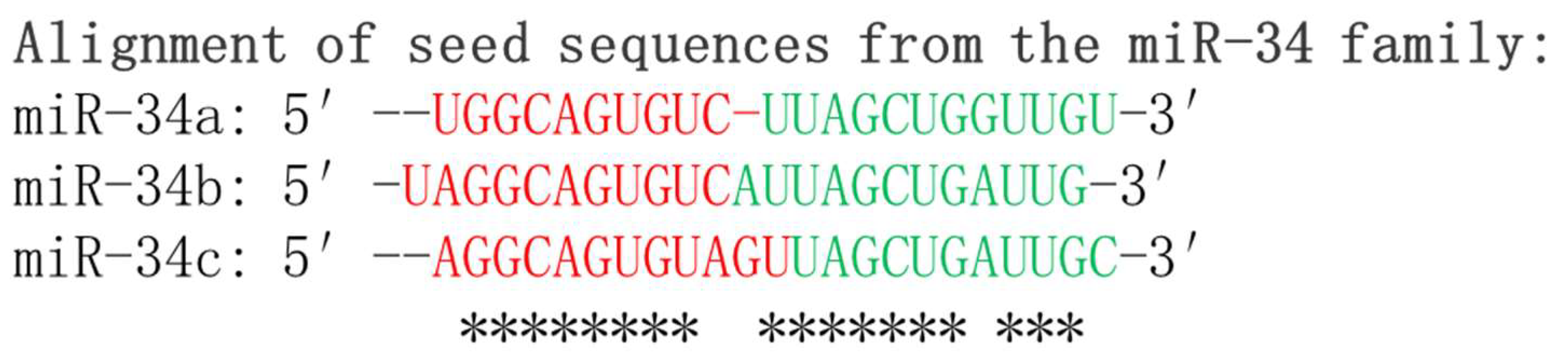
Figure 1. Sequence alignment of mature miR-34 family members: miR-34a, miR-34b, and miR-34c. The alignment showcases the highly conserved regions of these microRNAs, highlighting their functional importance. The seed sequences which are critical for target recognition by the miR-34 family are shown in red. * indicates conserved nucleotides of the seed sequences among family members.
1.2. Regulation of miR-34 Expression
The regulation of miR-34 expression involves multiple mechanisms that contribute to its dysregulation in cancer. Transcriptional regulation by various factors plays a crucial role in controlling miR-34 levels. The tumor suppressor protein p53 directly binds to the miR-34 gene promoter, activating its transcription. Similarly, other transcription factors, such as Elk-1, STAT3, Snail, Slug, ZEB1, and ZEB2, have been implicated in the regulation of miR-34 [2].
The dysregulation of EMT-TFs contributes to the acquisition of mesenchymal characteristics by cancer cells, resulting in their increased invasiveness and migratory capacity. EMT-TFs recruit chromatin-modifying enzymes, such as histone deacetylases and DNA methyltransferases (DNMTs), to the promoter regions of miR-34 genes. This recruitment leads to histone deacetylation and DNA methylation, causing a repressive chromatin state and inhibition of the miR-34 transcription [2].
Epigenetic modifications, including CpG island methylation and histone deacetylation, further contribute to the dysregulation of miR-34 in cancers. The methylation of CpG islands in the promoter regions of miR-34 genes results in gene silencing, while histone deacetylation leads to a more compact chromatin structure that hinders transcriptional activity [8]. These epigenetic modifications are commonly found in various cancer types, such as lung [9][10], multiple myeloma [11], kidney [12][13], gastric [14], breast [15][16], colorectal [17], hepatocellular [18], prostate [19], and ovarian [20][21] cancers.
Understanding the intricate regulatory networks involving transcriptional factors, EMT-TFs, and epigenetic modifications provides valuable insights into the molecular processes underlying cancer progression. Targeted therapies aimed at restoring miR-34 expression and inhibiting EMT-associated metastasis may hold promise for future therapeutic interventions in cancer treatment.
Given the tumor-suppressive function of miR-34s, correlations between the expression of miR-34s and patient survival were interrogated using a Cox proportional hazards regression and a Kaplan–Meier survival analysis (https://kmplot.com/analysis/index.php?p=service&cancer=pancancer_mirna) (access date: 24 April 2023) [22]. In multiple distinct cancer types, the high expression of miR-34s, such as miR-34a (Figure 2B, p = 0.0021), miR-34b (Figure 2F, p = 0.0001), and miR-34c, is correlated with longer survival of patients with liver hepatocellular carcinoma (HCC). For example, Chen et al. reported that patients with a lower level of serum exosomal miR-34a showed worse overall survival than patients with its high expression, implying it is a noninvasive marker for the diagnosis and prognosis of HCC [23].
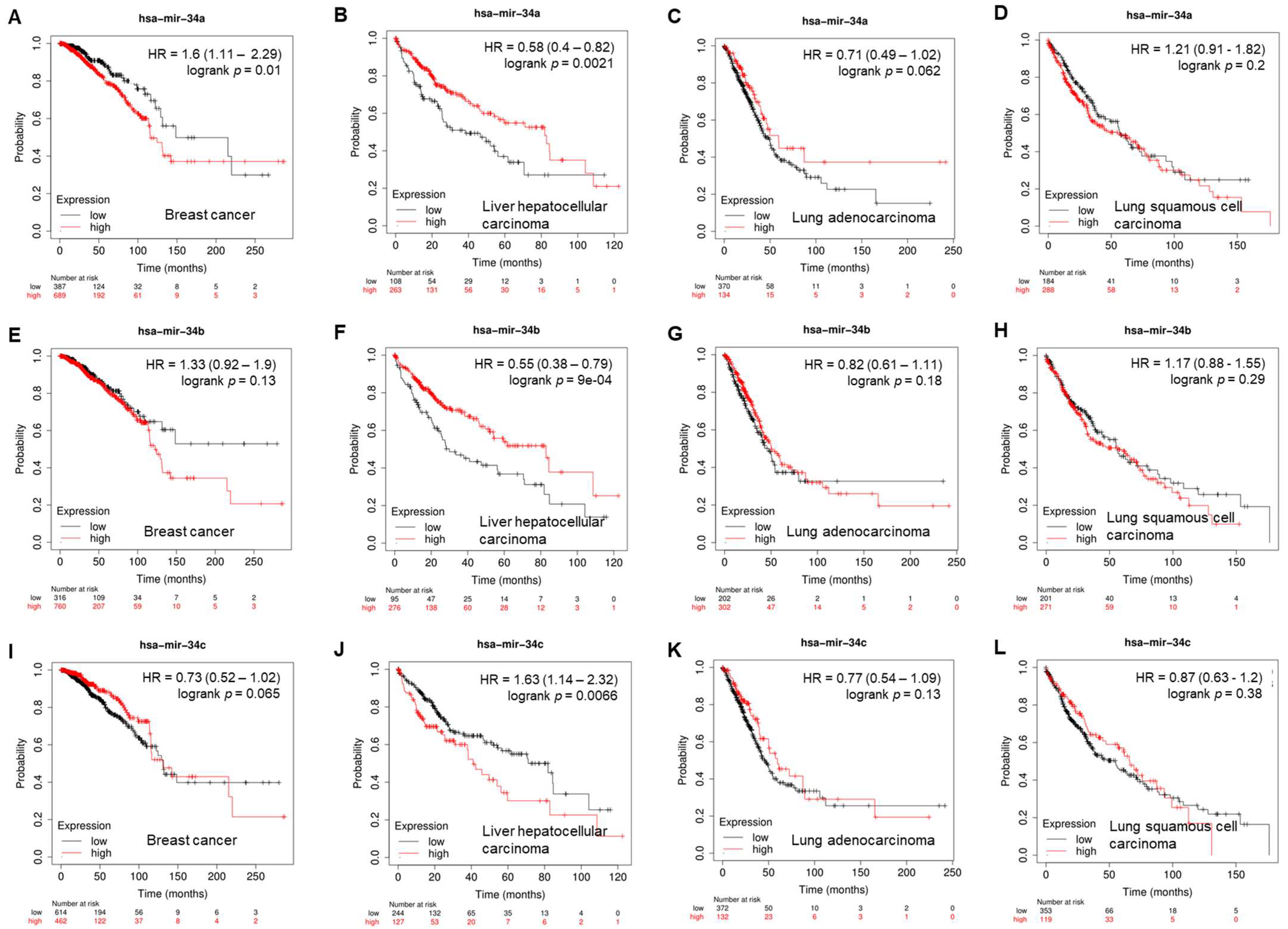
Figure 2. Correlation of overall survival with expression of miR-34a (A–D), miR-34b (E–H), and miR-34c (I–L) in breast cancer, liver hepatocellular carcinoma, lung adenocarcinoma, and lung squamous-cell carcinoma.
2. Exploring MicroRNA-34 Family Members in Drug Resistance
Resistance to conventional chemotherapy remains a significant challenge in cancer treatment. Therapy resistance leads to cancer relapse and poor patient outcomes. Numerous studies have demonstrated that altered levels of circulating miR-34s or tumor-specific miR-34 expressions are associated with poor responses to chemotherapy [24][25][26]. In patients with osteosarcoma, serum levels of miR-34a correlated with chemotherapy resistance, metastasis, recurrence, overall survival, and prognosis [27][28]. In colorectal cancers with inactive p53, targeting the miR-34a/LRPPRC/MDR1 axis has shown promise in overcoming resistance to chemotherapeutic agents such as gossypol-acetic acid and 5-fluorouracil (5-FU) [29]. The ability of miR-34a to suppress cancer stem cell renewal has been linked to a reduction in gemcitabine resistance in pancreatic cancer [30]. Furthermore, miR-34a has the ability to reverse multidrug resistance (MDR) to various chemotherapeutic agents including 5-FU, cisplatin (DDP), oxaliplatin, and epirubicin (EPI) in gastric cancer cells [31]. In bladder cancer, exosomal lncRNA LINC00355 derived from cancer-associated fibroblasts promotes resistance to DDP by targeting the miR-34b-5p/ABCB1 axis [32]. miR-34b expression has been reported to enhance paclitaxel chemosensitivity in endometrial cancer cells [33]. miR-34b also suppresses MDR to multiple drugs (paclitaxel, pirarubicin, EPI, hydrochloride, adriamycin, and DDP) in bladder cancer by regulating the expression of CCND2 and P2RY1 [34].
In the case of miR-34c, its role in drug resistance is complex and context-dependent. It has been reported to protect lung cancer cells from paclitaxel-induced apoptosis by regulating the expression of Bmf (Bcl-2-modifying factor) [35]. However, decreased miR-34c expression has also been observed in patients with NSCLC who exhibited a poor response to chemotherapy and increased metastasis. miR-34c overexpression sensitized NSCLC cells to paclitaxel and DDP both in vitro and in vivo [36]. miR-34c also inhibits MDR to paclitaxel and DDP in gastric cancer cells [37]. In ovarian cancer, the miR-34c/SOX9 axis regulates DDP-based drug resistance, and the miR-34c-AREG-EGFR-ERK pathway inhibits amphiregulin-induced cancer stemness and drug resistance [38][39].
Understanding the intricate roles of miR-34s in mediating drug resistance provides valuable insights for developing strategies to overcome treatment limitations and improve patient outcomes. Harnessing the potential of miR-34s as therapeutic targets or predictive biomarkers may lead to the development of personalized treatment approaches and the effective management of drug resistance in various cancer types.
Overcoming Chemoresistance by Targeting Cancer Stem Cells
Chemoresistance, a common hurdle in cancer treatment, has been attributed to the presence of a small population of cancer stem cells (CSCs) that possess self-renewal and differentiation capabilities [40]. These CSCs are thought to be responsible for tumor initiation, progression, and relapse. Due to its ability to suppress CSC activity, targeting CSCs using miR-34-based therapies has emerged as a promising approach to overcoming chemoresistance and improving treatment outcomes [41]. In taxane-resistant prostate cancer, the codelivery of DTX, a chemotherapy drug, and rub one (RUB), an activator of miR-34a specifically targeting CSCs, showed that the combination therapy effectively suppressed CSC-related markers, reduced the CSC population, and enhanced the therapeutic response compared to DTX alone [40][42]. Furthermore, miR-34 mimics have been shown to sensitize CSCs to chemotherapy alone. A combination of miR-34a mimics and doxorubicin, a commonly used chemotherapy drug used in breast cancer, synergistically inhibited breast cancer CSC properties and significantly reduced tumor growth compared to either treatment alone by sensitizing the CSCs to doxorubicin-induced apoptosis, thereby overcoming chemoresistance [43][44].
In addition to targeting CSCs directly, miR-34-based therapies have been explored for modulating the tumor microenvironment to disrupt CSC-related signaling pathways. For instance, miR-34a mimic delivery using nanocarriers was found to suppress the expression of CSC-associated factors and promote the differentiation of CSCs in hepatocellular carcinoma. This resulted in reduced tumor growth and enhanced chemosensitivity [45]. Moreover, the combination of miR-34 mimics with natural compounds known for their anticancer effects has shown promise in targeting CSCs. For example, the codelivery of miR-34a mimics with curcumin, a natural compound derived from turmeric, demonstrated synergistic effects in inhibiting CSC properties and suppressing tumor growth in colorectal cancer models [46]. Collectively, these studies highlight the potential of miR-34-based therapies in targeting CSCs and overcoming chemoresistance. By specifically inhibiting CSCs and their associated signaling pathways, miR-34 mimics hold promise for improving treatment outcomes and address the challenges posed by CSC-mediated resistance in cancer therapy.
This entry is adapted from the peer-reviewed paper 10.3390/cancers15194723
References
- Hermeking, H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010, 17, 193–199.
- Agostini, M.; Knight, R.A. miR-34: From bench to bedside. Oncotarget 2014, 5, 872–881.
- Rokavec, M.; Li, H.; Jiang, L.; Hermeking, H. The p53/miR-34 axis in development and disease. J. Mol. Cell Biol. 2014, 6, 214–230.
- Maroof, H.; Salajegheh, A.; Smith, R.A.; Lam, A.K. Role of microRNA-34 family in cancer with particular reference to cancer angiogenesis. Exp. Mol. Pathol. 2014, 97, 298–304.
- Pillai, R.S.; Bhattacharyya, S.N.; Filipowicz, W. Repression of protein synthesis by miRNAs: How many mechanisms? Trends Cell Biol. 2007, 17, 118–126.
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233.
- Shukla, G.C.; Singh, J.; Barik, S. MicroRNAs: Processing, Maturation, Target Recognition and Regulatory Functions. Mol. Cell Pharmacol. 2011, 3, 83–92.
- Wang, C.; Jia, Q.; Guo, X.; Li, K.; Chen, W.; Shen, Q.; Xu, C.; Fu, Y. microRNA-34 family: From mechanism to potential applications. Int. J. Biochem. Cell Biol. 2022, 144, 106168.
- Bommer, G.T.; Gerin, I.; Feng, Y.; Kaczorowski, A.J.; Kuick, R.; Love, R.E.; Zhai, Y.; Giordano, T.J.; Qin, Z.S. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr. Biol. 2007, 17, 1298–1307.
- Zhao, K.; Cheng, J.; Chen, B.; Liu, Q.; Xu, D.; Zhang, Y. Circulating microRNA-34 family low expression correlates with poor prognosis in patients with non-small cell lung cancer. J. Thorac. Dis. 2017, 9, 3735–3746.
- Yamazaki, H.; Chijiwa, T.; Inoue, Y.; Abe, Y.; Suemizu, H.; Kawai, K.; Wakui, M.; Furukawa, D.; Mukai, M.; Kuwao, S. Overexpression of the miR-34 family suppresses invasive growth of malignant melanoma with the wild-type p53 gene. Exp. Ther. Med. 2012, 3, 793–796.
- Beresneva, E.V.; Rykov, S.V.; Hodyrev, D.S.; Pronina, I.V.; Ermilova, V.D.; Kazubskaia, T.P.; Braga, E.A.; Loginov, V.I. Methylation profile of group of miRNA genes in clear cell renal cell carcinoma; involvement in cancer progression. Genetika 2013, 49, 366–375.
- Fujino, T.; Yokosuka, A.; Higurashi, H.; Yokokawa, R.; Sakurai, R.; Harashima, W.; Miki, Y.; Fujiwara, Y.; Mimaki, Y.; Hayakawa, M. AU-1 from Agavaceae plants causes transient increase in p21/Cip1 expression in renal adenocarcinoma ACHN cells in an miR-34-dependent manner. J. Nat. Med. 2017, 71, 36–43.
- Ji, Q.; Hao, X.; Meng, Y.; Zhang, M.; Desano, J.; Fan, D.; Xu, L. Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheres. BMC Cancer 2008, 8, 266.
- Imani, S.; Wei, C.; Cheng, J.; Khan, M.A.; Fu, S.; Yang, L.; Tania, M.; Zhang, X.; Xiao, X.; Zhang, X.; et al. MicroRNA-34a targets epithelial to mesenchymal transition-inducing transcription factors (EMT-TFs) and inhibits breast cancer cell migration and invasion. Oncotarget 2017, 8, 21362–21379.
- Kaboli, P.J.; Rahmat, A.; Ismail, P.; Ling, K.H. MicroRNA-based therapy and breast cancer: A comprehensive review of novel therapeutic strategies from diagnosis to treatment. Pharmacol. Res. 2015, 97, 104–121.
- Toyota, M.; Suzuki, H.; Sasaki, Y.; Maruyama, R.; Imai, K.; Shinomura, Y.; Tokino, T. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008, 68, 4123–4132.
- Liu, C.J.; Ma, X.W.; Zhang, X.J.; Shen, S.Q. Erratum: Pri-miR-34b/c rs4938723 polymorphism is associated with hepatocellular carcinoma risk: A case-control study in a Chinese population. Int. J. Mol. Epidemiol. Genet. 2017, 8, 59.
- Wang, L.; Yu, J.; Xu, J.; Zheng, C.; Li, X.; Du, J. The analysis of microRNA-34 family expression in human cancer studies comparing cancer tissues with corresponding pericarcinous tissues. Gene 2015, 554, 1–8.
- Welponer, H.; Tsibulak, I.; Wieser, V.; Degasper, C.; Shivalingaiah, G.; Wenzel, S.; Sprung, S.; Marth, C.; Hackl, H.; Fiegl, H. The miR-34 family and its clinical significance in ovarian cancer. J. Cancer 2020, 11, 1446–1456.
- Corney, D.C.; Hwang, C.I.; Matoso, A.; Vogt, M.; Flesken-Nikitin, A.; Godwin, A.K.; Kamat, A.A.; Sood, A.K.; Ellenson, L.H.; Hermeking, H. Frequent downregulation of miR-34 family in human ovarian cancers. Clin. Cancer Res. 2010, 16, 1119–1128.
- Nagy, A.; Munkacsy, G.; Gyorffy, B. Pancancer survival analysis of cancer hallmark genes. Sci. Rep. 2021, 11, 6047.
- Chen, S.; Mao, Y.; Chen, W.; Liu, C.; Wu, H.; Zhang, J.; Wang, S.; Wang, C.; Lin, Y.; Lv, Y. Serum exosomal miR-34a as a potential biomarker for the diagnosis and prognostic of hepatocellular carcinoma. J. Cancer 2022, 13, 1410–1417.
- Hu, Y.; Qiu, Y.; Yagüe, E.; Ji, W.; Liu, J.; Zhang, J. miRNA-205 targets VEGFA and FGF2 and regulates resistance to chemotherapeutics in breast cancer. Cell Death Dis. 2016, 7, e2291.
- Teoh, S.L.; Das, S. The Role of MicroRNAs in Diagnosis, Prognosis, Metastasis and Resistant Cases in Breast Cancer. Curr. Pharm. Des. 2017, 23, 1845–1859.
- Lian, H.; Zhou, Y.; Sun, Z.; Liu, K. MicroRNA34a is associated with chemotherapy resistance, metastasis, recurrence, survival, and prognosis in patient with osteosarcoma. Medicine 2022, 101, e30722.
- Li, S.; Wei, X.; He, J.; Cao, Q.; Du, D.; Zhan, X.; Zeng, Y.; Yuan, S.; Sun, L. The comprehensive landscape of miR-34a in cancer research. Cancer Metastasis. Rev. 2021, 40, 925–948.
- Yang, Y.; Yuan, H.; Zhao, L.; Guo, S.; Hu, S.; Tian, M.; Nie, Y.; Yu, J.; Zhou, C.; Niu, J. Targeting the miR-34a/LRPPRC/MDR1 axis collapse the chemoresistance in P53 inactive colorectal cancer. Cell Death Differ. 2022, 29, 2177–2189.
- Pan, Y.; Li, K.; Tao, X.; Zhao, Y.; Chen, Q.; Li, N.; Liu, J.; Go, V.L.W.; Guo, J.; Gao, G. MicroRNA-34a Alleviates Gemcitabine Resistance in Pancreatic Cancer by Repression of Cancer Stem Cell Renewal. Pancreas 2021, 50, 1260–1266.
- Deng, X.J.; Zheng, H.L.; Ke, X.Q.; Deng, M.; Ma, Z.Z.; Zhu, Y.; Cui, Y.Y. Hsa-miR-34a-5p reverses multidrug resistance in gastric cancer cells by targeting the 3’-UTR of SIRT1 and inhibiting its expression. Cell Signal 2021, 84, 110016.
- Luo, G.; Zhang, Y.; Wu, Z.; Zhang, L.; Liang, C.; Chen, X. Exosomal LINC00355 derived from cancer-associated fibroblasts promotes bladder cancer cell resistance to cisplatin by regulating miR-34b-5p/ABCB1 axis. Acta Biochim. Biophys. Sin. 2021, 53, 558–566.
- Yanokura, M.; Banno, K.; Aoki, D. MicroRNA-34b expression enhances chemosensitivity of endometrial cancer cells to paclitaxel. Int. J. Oncol. 2020, 57, 1145–1156.
- Tan, Y.; Zhang, T.; Zhou, L.; Liu, S.; Liang, C. MiR-34b-3p Represses the Multidrug-Chemoresistance of Bladder Cancer Cells by Regulating the CCND2 and P2RY1 Genes. Med. Sci. Monit. 2019, 25, 1323–1335.
- Catuogno, S.; Cerchia, L.; Romano, G.; Pognonec, P.; Condorelli, G.; de Franciscis, V. miR-34c may protect lung cancer cells from paclitaxel-induced apoptosis. Oncogene 2013, 32, 341–351.
- Yang, L.Z.; Lei, C.C.; Zhao, Y.P.; Sun, H.W.; Yu, Q.H.; Yang, E.J.; Zhan, X. MicroRNA-34c-3p target inhibiting NOTCH1 suppresses chemosensitivity and metastasis of non-small cell lung cancer. J. Int. Med. Res. 2020, 48, 300060520904847.
- Zheng, H.; Wang, J.J.; Yang, X.R.; Yu, Y.L. Upregulation of miR-34c after silencing E2F transcription factor 1 inhibits paclitaxel combined with cisplatin resistance in gastric cancer cells. World J. Gastroenterol. 2020, 26, 499–513.
- Xiao, S.; Li, Y.; Pan, Q.; Ye, M.; He, S.; Tian, Q.; Xue, M. MiR-34c/SOX9 axis regulates the chemoresistance of ovarian cancer cell to cisplatin-based chemotherapy. J. Cell Biochem. 2019, 120, 2940–2953.
- Tung, S.L.; Huang, W.C.; Hsu, F.C.; Yang, Z.P.; Jang, T.H.; Chang, J.W.; Chuang, C.M.; Lai, C.R.; Wang, L.H. miRNA-34c-5p inhibits amphiregulin-induced ovarian cancer stemness and drug resistance via downregulation of the AREG-EGFR-ERK pathway. Oncogenesis 2017, 6, e326.
- Yoshida, K.; Yamamoto, Y.; Ochiya, T. miRNA signaling networks in cancer stem cells. Regen. Ther. 2021, 17, 1–7.
- Li, W.J.; Wang, Y.; Liu, R.; Kasinski, A.L.; Shen, H.; Slack, F.J.; Tang, D.G. MicroRNA-34a: Potent Tumor Suppressor, Cancer Stem Cell Inhibitor, and Potential Anticancer Therapeutic. Front. Cell Dev. Biol. 2021, 9, 640587.
- Lin, F.; Wen, D.; Wang, X.; Mahato, R.I. Dual responsive micelles capable of modulating miRNA-34a to combat taxane resistance in prostate cancer. Biomaterials 2019, 192, 95–108.
- Vares, G.; Ahire, V.; Sunada, S.; Ho Kim, E.; Sai, S.; Chevalier, F.; Romeo, P.H.; Yamamoto, T.; Nakajima, T.; Saintigny, Y. A multimodal treatment of carbon ions irradiation, miRNA-34 and mTOR inhibitor specifically control high-grade chondrosarcoma cancer stem cells. Radiother. Oncol. 2020, 150, 253–261.
- Imani, S.; Wu, R.C.; Fu, J. MicroRNA-34 family in breast cancer: From research to therapeutic potential. J. Cancer 2018, 9, 3765–3775.
- Daverey, A.; Brown, K.M.; Kidambi, S. Breast Cancer/Stromal Cells Coculture on Polyelectrolyte Films Emulates Tumor Stages and miRNA Profiles of Clinical Samples. Langmuir 2015, 31, 9991–10001.
- Shi, L.; Wang, Z.; Geng, X.; Zhang, Y.; Xue, Z. Exosomal miRNA-34 from cancer-associated fibroblasts inhibits growth and invasion of gastric cancer cells in vitro and in vivo. Aging 2020, 12, 8549–8564.
- Kalfert, D.; Ludvikova, M.; Pesta, M.; Ludvik, J.; Dostalova, L.; Kholova, I. Multifunctional Roles of miR-34a in Cancer: A Review with the Emphasis on Head and Neck Squamous Cell Carcinoma and Thyroid Cancer with Clinical Implications. Diagnostics 2020, 10, 563.
This entry is offline, you can click here to edit this entry!
