Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Microbial secondary metabolites are low-molecular-weight compounds synthesized by microorganisms after the growth phase. Secondary metabolites are not directly involved in microbial growth. Solid-state fermentation (SSF) is a process whereby microorganisms grow in the absence of free water or with low water content. It has been used since ancient times to obtain fermented foods such as koji, bread, and cheeses.
- agro-industrial residue
- microbial secondary metabolite
- biotransformation
1. Introduction
Microbial secondary metabolites are low-molecular-weight compounds synthesized by microorganisms after the growth phase. Secondary metabolites are not directly involved in microbial growth. Still, they play a significant role in competition, antagonism, and self-defense mechanisms [1]. These compounds have a variety of chemical structures, and many have biological activities such as antimicrobial, antiviral, antioxidant, antitumor, vasodilator, vasoconstrictor, diuretic, and laxative activities, among others [2]. Utilizing microbial cultures for secondary metabolite production has proven to be an effective strategy for reducing production costs and carbon footprints. Moreover, microbial production also contributes to preserving plant biodiversity [3] since microorganisms can produce some plant secondary metabolites through the biotransformation of precursors. Microbial production then allows humans to avoid the disadvantages of plant extraction, like low yield, which leads to plant over-exploitation.
Production of microbial secondary metabolites is influenced by diverse culture conditions such as the composition of culture media, the carbon/nitrogen ratio, salinity, the presence of metal ions, temperature, pH, and oxygen concentration [4]. The fermentation system significantly influences the production of secondary metabolites. They are mainly produced by submerged fermentation (SmF) due to the ease of measuring, monitoring, and controlling process variables. Nonetheless, solid-state fermentation (SSF) is recognized to imitate the natural environment for the growth of microorganisms, representing an advantage for obtaining some metabolites [5].
SSF is a process whereby microorganisms grow in the absence of free water or with low water content [6]. It has been used since ancient times to obtain fermented foods such as koji, bread, and cheeses. Even in the last century, SSF has produced compounds for the food, pharmaceutical, textile, biochemical, and energy industries [7]. This innovative culture system has produced a wide range of secondary metabolites, including bioactive compounds, enzymes, polyphenols, food additives, and others [8][9][10].
There are two types of SSF. The first is the most common and uses a natural solid material acting as substrate and support. The second uses an inert support impregnated with a nutritive culture medium. In both systems, advantages of SSF over SmF have been reported, such as higher yield in the production of enzymes and secondary metabolites. Furthermore, some microorganisms only produce certain enzymes or secondary metabolites in SSF, even though they can grow well in SmF [11]. Therefore, different authors have used SSF to produce pigments, antibiotics, statins, biosurfactants, phenolic compounds, and other secondary metabolites. Table 1 shows some examples of secondary metabolites that SSF can obtain.
Table 1. Examples of secondary metabolites produced by SSF.
| Metabolite and Its Potential Application | Microorganism | Solid Support | Yield (mg/g SS) | Reference |
|---|---|---|---|---|
| Pigments | ||||
| Monascin, ankaflavin, rubropunctatin, monascorubrin, rubropunctamin, monascorubramine, etc. | Monascus sp. | Rice | 49.65 * | [12][13] |
| Antibiotics | ||||
| Penicillin | Penicillium chrysogenum | Sugarcane bagasse | 7–8 | [14][15] |
| Cephalosporin C | Acremonium chrysogenum | Sugarcane bagasse | 3.2 | [16][17] |
| Paromomycin | Streptomyces rimosus | Corn bran | 2.2 | [18] |
| Neomycin | Streptomyces fradiae | Nylon sponge | n.d. | [19] |
| Rifamycin B | Nocardia mediterranei | Sunflower oil cake | 9.87 | [20] |
| Antifungal | ||||
| Sclerotiorin | Penicillium sclerotiorum | Rice | n.d. | [21] |
| Griseofulvin | Penicillium griseofulvum | Rice bran | 9–10 | [22] |
| Natamycin | Streptomyces gilvosporeus | Wheat bran, rapeseed cake, and rice hull | 9.62 | [23] |
| Statins | ||||
| Lovastatin | Aspergillus terreus | Glucose and lactose | 19.95–25 | [24][25][26] |
| Compactin (mevastatin) | Penicillium brevicompactum | Soybean meal | 1.406 | [27] |
| Monacolin K | Monascus ruber | Millet | 19.81 | [28] |
| Rice and bran | 14.53 | [29] | ||
| Biosurfactants | ||||
| Surfactin | Bacillus subtilis | Olive cake flour | 30.67 | [30] |
| Bacillus amyloliquefaciens | Soybean flour | 15.03 | [31] | |
| Iturin | Bacillus subtilis | Defatted soybean meal, wheat bran and ricehusk | 5.58 | [32] |
| Rhamnolipids | Pseudomonas aeruginosa | Polyurethane foam | 39.8 ** | [33] |
| Soybean meal | 19.68 | [34] | ||
| Sophorolipids | Starmerella bombicola | Polyurethane foam | 211 *** | [35] |
| Wheat straw | 195 | [36] | ||
| Phenolics | ||||
| Vainillin | Enterobacter hormaechei | Sugarcane bagasse | 4.76 | [37] |
| Enterobacter hormaechei | Pomegranate peels | 0.462 | [38] | |
| Streptomyces sannanensis | Wheat straw | 2.74 | [39] | |
| Gallic acid | Aspergillus niger | Black plum seed | 14.5 | [40] |
| Hispidin | Phellinus linteus | Brown rice and pearl barley | 0.375 | [41] |
| Immunosuppressants | ||||
| Mycophenolic acid | Penicillium brevicompactum | Parmal rice | 4.5 | [42] |
| Cyclosporin A | Tolypocladium inflatum | Wheat bran flour and coconut oil cake | 6.48 | [43] |
| Phytohormones | ||||
| Gibberellic acid | Gibberella fujikuroi | Amberlite IRA-900 | n.d. | [44] |
| Alkaloids | ||||
| Ergotamine | Claviceps purpurea | Rice | ≈0.015 | [45] |
* Optical density unit (ODU)/g SS, ** g/L, *** mg/g substrate, n.d. = not determined.
Fungal pigments are probably the first secondary metabolites commercially produced by SSF. Fungi of the genus Monascus have been cultivated on cooked rice in Asian countries since ancient times to make a red colorant known as “Anka,” or “red mold rice,” which is used as a food ingredient [46]. Monascus sp. produces orange, red, and yellow pigments in these conditions. These pigments are a mixture of secondary metabolites such as monascin, ankaflavin, rubropunctatin, monascorubrin, rubropunctamin, and monascorubramine, among others [12][13].
Antibiotics are one of the best-known groups of microbial secondary metabolites. Antibiotics act against other microorganisms affecting cellular processes such as DNA replication, transcription, cell wall synthesis, and cell membrane disruption [47]. Antibiotics are produced commercially by SmF. However, SSF is an alternative system with advantages for large-scale production, such as higher yields in shorter periods [11]. For this reason, various authors have investigated the production of antibiotics such as penicillin [14][15], cephalosporin C [16][17], paramomycin [18], neomycin [19], and rifamycin [20] by SSF.
On the other hand, statins are a group of drugs that lower blood cholesterol levels, decreasing the risk of heart attack or stroke. Filamentous fungi produce natural statins (lovastatin, compactin, and monacolin K). Notably, the industrial production of lovastatin is mainly carried out by SmF using Aspergillus terreus [48]. However, several authors have described the advantages of SSF for lovastatin production compared to SmF. For example, Baños et al. [24] reported 30 times higher lovastatin production by A. terreus TUB F-514 in SSF than in SmF. In addition, the specific production was 14 times higher in SSF. Therefore, the Indian biotech company Biocon Ltd. developed a method to produce lovastatin in SSF using the Plafractor bioreactor [49]. Lovastatin produced by SSF received FDA approval for sale in the United States in 2001.
Other molecules produced at the end of the exponential growth phase of certain bacteria, yeasts, and molds are biosurfactants, which reduce surface and interfacial tension due to their amphiphilic nature. These molecules are involved in cell development, biofilm formation, osmotic pressure regulation, and hydrophobic substance assimilation [50]. They are produced by certain microorganisms, mainly by SmF. Still, during their manufacture, a large amount of foam is produced, increasing the risk of contamination and reducing productivity. Conversely, SSF eliminates the foaming problem and reduces energy and water consumption during production [34]. For this reason, different researchers have investigated SSF to produce biosurfactants such as sophorolipids [35][36], rhamnolipids [33][34], surfactin [30][31], and iturin [32], among others.
Some secondary metabolites obtained from plants cannot be produced directly by microorganisms. However, some microorganisms can biotransform the chemical precursors of those plants’ secondary metabolites. For example, vanillin is an essential flavoring agent in the food industry, traditionally extracted from vanilla pods. Several bacteria of the genera Amycolatopsis, Streptomyces, Pseudomonas, Delftia, and Enterobacter can catalyze the conversion of ferulic acid to vanillin. Some authors have investigated the use of ferulic acid-rich agro-industrial by-products such as sugarcane bagasse [37], pomegranate peels [38], and wheat straw [39] to produce biovanillin through SSF.
2. Main Biosynthetic Pathways of Secondary Metabolism
Secondary metabolites are not essential for organisms’ growth, development, reproduction, or energy production. Therefore, these compounds are not produced by all microbial species. Secondary metabolites are synthesized from primary metabolites, such as acetyl-coenzyme A and amino acids, through secondary metabolic pathways [51].
Two metabolic pathways synthesize the precursors of phenolic compounds: the shikimic acid and malonic acid pathways. The shikimic acid pathway is most important in plants. The malonic acid pathway is an essential source of phenolics in fungi and bacteria but is less significant in plants [52]. Terpenoid precursors can be synthesized by two metabolic pathways: the mevalonic acid pathway or the methylerythritol phosphate pathway. The mevalonic acid pathway is present in plants, animals, yeast, fungi, archaea, and some eubacteria. The methylerythritol phosphate pathway is present in most bacteria, cyanobacteria, and plant plastids [53]. Primary and secondary metabolic pathways are interrelated, as shown in Figure 1. The regulation of secondary metabolism is a complex process. Therefore, in order to improve the production of secondary metabolites by microorganisms, it is essential to understand the metabolic pathways involved. The main metabolic pathways related to the production of secondary metabolites in microorganisms are described below.
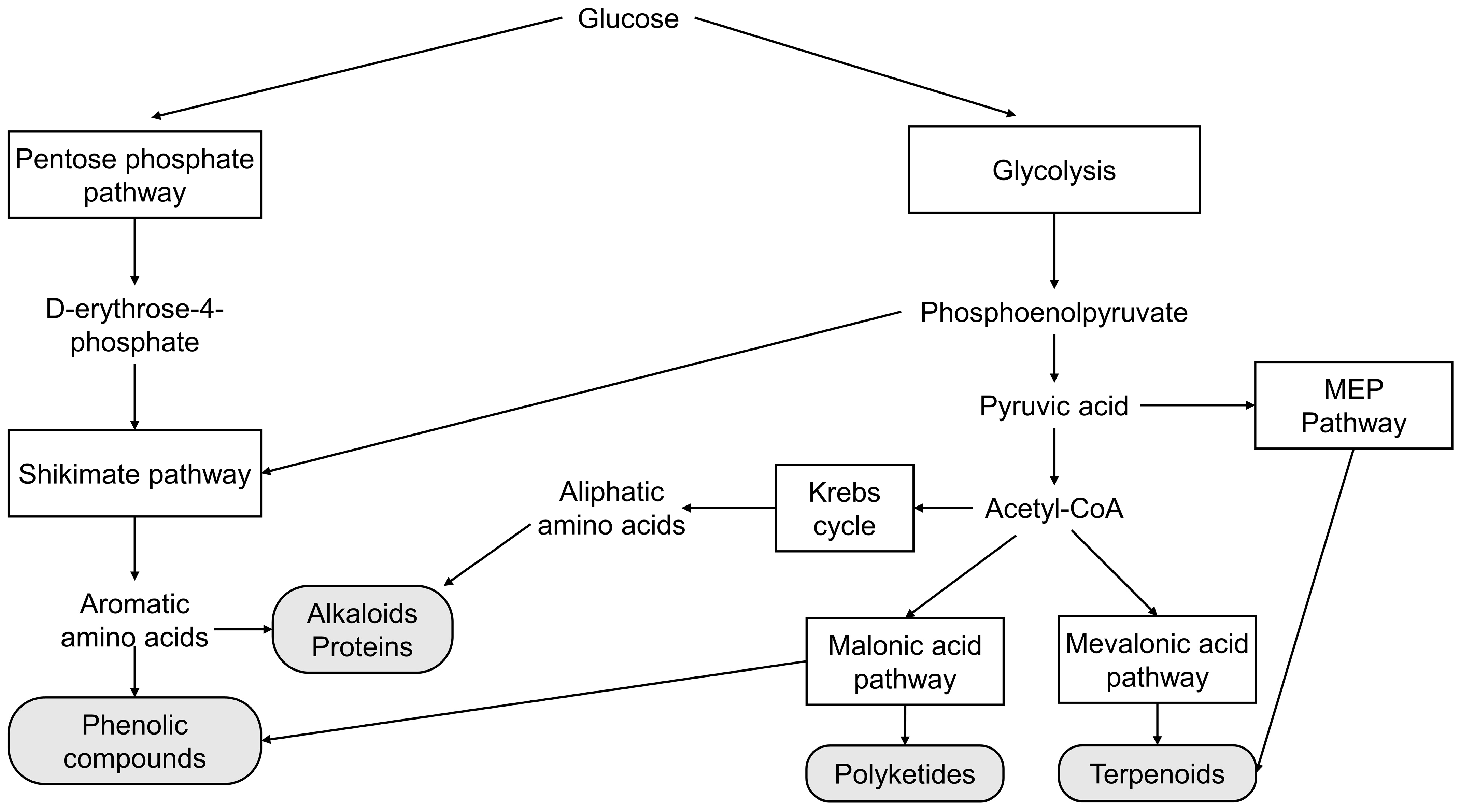
Figure 1. Schematic diagram of the main metabolic pathways involved in the production of secondary metabolites by microorganisms.
2.1. Shikimate Pathway
The shikimate pathway provides precursors for aromatic molecules in bacteria, fungi, and plants but not in animals. This pathway provides the aromatic amino acids (L-phenylalanine, L-tyrosine, and L-tryptophan) necessary for protein synthesis. Aromatic amino acids also serve as precursors for secondary metabolites, such as phenolic compounds and some alkaloids [54].
The shikimate pathway consists of seven enzymatic reactions: First, phosphoenolpyruvate (an intermediate metabolite in the Embden–Meyerhof–Parnas pathway) and D-erythrose-4-phosphate (an intermediate metabolite in the pentose phosphate pathway) are converted to 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) by DAHP synthase. DAHP is then converted to shikimic acid by 3-dehydroquinate (DHQ) synthase, DHQ dehydratase, and shikimate dehydrogenase. Last, shikimic acid is converted to chorismic acid by shikimate kinase, 5-enolpyruvylshikimate 3-phosphate synthase, and chorismic acid synthase. The final product of the shikimate pathway (chorismic acid) can be further transformed into aromatic amino acids by a single reaction. [55]. The phenylpropanoid pathway transforms L-tyrosine and L-phenylalanine into various phenolic compounds in higher plants. No evidence exists of complete phenylpropanoid metabolism in organisms other than land plants. However, some homologous enzymes of this pathway have been found in some bacteria and fungi [56].
The shikimate pathway has different metabolic branches in different microorganisms that lead to the formation of diverse secondary metabolites, such as shikimic acid, gallic acid pyrogallol, chlorogenic acid, and catechol [57]. For example, shikimic acid is an intermediate compound of this pathway. It has a highly functionalized, six-carbon ring with three chiral carbons and a carboxylic acid functional group. Therefore, it is widely used to synthesize valuable products such as the antiviral drug oseltamivir (Tamiflu®) [58].
2.2. Malonic Acid Pathway
Most of the phenolic compounds in higher plants are produced by the shikimate pathway, whereas in bacteria and fungi, the phenolic compounds are also synthesized by the malonic acid pathway [59].
The key enzyme tyrosine ammonia lyase deaminates tyrosine into p-coumaric acid in the malonic acid pathway. It is functionalized into p-coumaroyl-CoA by 4-coumaroyl CoA ligase. Then it reacts with three molecules of malonyl-CoA to give chalcone by chalcone synthase. The obtained tetraoxychalcone is transformed into flavanone–naringenin, which serves as a precursor to other flavonoids [60].
2.3. Mevalonic Acid Pathway (MVA)
The MVA pathway is present in most eukaryotes, archaea, and some bacteria. This pathway begins with the condensation of two acetyl-CoA molecules. It ends with the formation of isopentenyl-pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP), the precursors to terpenoid biosynthesis [61].
In the MVA pathway, 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) is produced from the sequential condensation of three molecules of acetyl-CoA catalyzed by acetoacetyl-CoA thiolase, and HMG-CoA synthase HMG-CoA is then converted to mevalonic acid by HMG-CoA reductase (HMGCR). Mevalonic acid is sequentially phosphorylated to 5-phosphomevalonate and 5-diphosphomevalonate and decarboxylated to generate IPP by the enzymes mevalonate kinase, 5-phosphomevalonate kinase, and 5-diphosphomevalonate decarboxylase. Finally, DMAPP is produced from IPP by a reversible reaction catalyzed by IPP/DMAPP isomerase [62].
HMGCR is the rate-limiting enzyme in the MVA pathway. There are two classes of HMGCR. Class I includes proteins of eukaryotic origin that are associated with the endoplasmic reticulum and are potentially inhibited by statins. Class II proteins of bacterial origin have low homology with class I HMGCRs (<20%). However, there is considerable similarity between the active sites of both classes of enzymes [63].
2.4. Methylerythritol-Phosphate (MEP) Pathway
The MEP pathway is an alternative route to MVA to produce terpenoid precursors (IPP and DMAPP). Most bacteria, cyanobacteria, and green algae exclusively use the MEP pathway. At the same time, plastid-bearing organisms have both pathways compartmentalized in the cytosol (MVA) and plastids (MEP) [64].
The MEP pathway initiates with the condensation between D-glyceraldehyde 3-phosphate and pyruvate to produce 1-deoxy-D-xylulose 5-phosphate (DXP), catalyzed by DXP synthase. DXP is then reductively isomerized to methylerythritol phosphate (MEP) by DXP reductoisomerase. Coupling between MEP and CTP is catalyzed by CDP-ME synthetase to produce methylerythritol cytidyl diphosphate (CDP-ME). CDP-ME is then phosphorylated to 4-diphosphocytidyl-2-C-methyl-D-erythritol-2-phosphate (CDP-MEP). CDP-MEP is cyclized to 2-C-methyl-D-erythritol-2,4-cyclodiphosphate (MEcPP). The opening of the cyclic pyrophosphate and the C3-reductive dehydration of MEcPP is catalyzed by 2-C-methyl-D-erythritol-2,4-cyclodiphosphate reductase to yield 4-hydroxy-3-methyl-butenyl 1-diphosphate (HMBPP). Finally, HMBPP is reduced to IPP and DMAPP by 4-hydroxyl-3-methyl-butenyl 1-diphosphate reductase [65].
Finally, it should be emphasized that although common molecular patterns and principles underlie life’s diverse forms, many differences in primary biosynthetic pathways exist. Many biosynthetic pathways are specific to certain groups of organisms. For instance, pheammonium lyase is a ubiquitous enzyme in fungi that catalyzes the deamination of L-Phe to trans-cinnamic acid. Conversely, only a few cinnamic and benzoic acid-derived metabolites have been described in prokaryotes [66].
From a practical perspective, the choice between using fungi or bacteria depends on the specific metabolite to be produced. Particular fungal or bacterial species exclusively synthesize some metabolites. Vancomycin, a glycopeptide antibiotic class, is produced by Amycolatopsis (formerly Streptomyces) species, such as A. orientalis or A. keratiniphila [67][68]. Although vancomycin is effective against methicillin-resistant Staphylococcus aureus infections [68], the emergence of vancomycin-resistant S. aureus has prompted the development of second-generation glycopeptide antibiotics. An example is the recently FDA-approved compound oritavancin. Although semi-synthetic, its production still relies on the in vivo production of vancomycin by Amycolatopsis species, and then the chassis is modified by incorporating a 4-(4-chlorophenyl) benzyl group through reductive alkylation [68].
Similarly, some secondary metabolites are exclusively synthesized by fungi. Beauvericin belongs to the cyclic hexadepsipeptide family and is produced via a non-ribosomal pathway utilizing beauvericin synthetase. It sequentially binds hydroxy isovaleric acid and N-methyl-phenylalanine molecules [69]. Certain entomopathogenic fungi generate Beauvericin, which exhibits diverse biological activities, including insecticidal, antimicrobial, and antitumor properties. Due to the intricate nature of its chemical synthesis, beauvericin production is predominantly accomplished via in vivo biosynthesis using specialized producer strains. Recently, Vásquez-Bonilla et al. [69] reported an enhancement in beauvericin production using solid-state cultures of Fusarium oxysporum AB2 compared to liquid cultures, increasing the yield from 0.8 mg/L to 65.3 mg/L. Moreover, they further improved yields by employing mixed cultures of F. oxysporum AB2 and Epicoccum nigrum TORT, producing 84.6 mg/L.
This entry is adapted from the peer-reviewed paper 10.3390/fermentation9090804
References
- Kumar, V.; Ahluwalia, V.; Saran, S.; Kumar, J.; Patel, A.K.; Singhania, R.R. Recent Developments on Solid-State Fermentation for Production of Microbial Secondary Metabolites: Challenges and Solutions. Bioresour. Technol. 2021, 323, 124566.
- Fouillaud, M.; Dufossé, L. Microbial Secondary Metabolism and Biotechnology. Microorganisms 2022, 10, 123.
- Mishra, S.; Sahu, P.K.; Agarwal, V.; Singh, N. Exploiting Endophytic Microbes as Micro-Factories for Plant Secondary Metabolite Production. Appl. Microbiol. Biotechnol. 2021, 105, 6579–6596.
- Pan, R.; Bai, X.; Chen, J.; Zhang, H.; Wang, H. Exploring Structural Diversity of Microbe Secondary Metabolites Using OSMAC Strategy: A Literature Review. Front. Microbiol. 2019, 10, 294.
- Rodríguez-Durán, L.V.; Michel, M.R.; Pichardo, A.; Aguilar-Zárate, P. Microbial Bioreactors for Secondary Metabolite Production. In Microbial Bioreactors for Industrial Molecules; Wiley: Hoboken, NJ, USA, 2023; pp. 275–296.
- Cerda-Cejudo, N.D.; Buenrostro-Figueroa, J.J.; Sepúlveda-Torre, L.; Torres-León, C.; Chávez-González, M.L.; Ascacio-Valdés, J.A.; Aguilar, C.N. Solid-State Fermentation for the Recovery of Phenolic Compounds from Agro-Wastes. Resources 2023, 12, 36.
- Soccol, C.R.; Costa, E.S.F.d.; Letti, L.A.J.; Karp, S.G.; Woiciechowski, A.L.; Vandenberghe, L.P.d.S. Recent Developments and Innovations in Solid State Fermentation. Biotechnol. Res. Innov. 2017, 1, 52–71.
- Barrios-González, J. Secondary Metabolites Production: Physiological Advantages in Solid-State Fermentation. In Current Developments in Biotechnology and Bioengineering; Elsevier: Amsterdam, The Netherlands, 2018; pp. 257–283.
- Jazmín Edith, M.-H.; Octavio, L.; Edna Madai, M.-H.; Esperanza, H.; Nicolás Óscar, S.-C. Coupling Energy-Production Processes: The Use of Residues from Bioethanol Production to Improve the Anaerobic Digestion of Corn Stover. Biomass Bioenergy 2019, 128, 105322.
- Sainz-Mellado, D.C.; Méndez-Hernández, J.E.; López-Miranda, J.; Páez-Lerma, J.B.; Aguilar, C.N.; Soto-Cruz, N.O. Gradually Supply of Isoamyl Alcohol Increases the Isoamyl Acetate Production in Solid-State Fermentation. Lett. Appl. Microbiol. 2023, 76, ovac061.
- Barrios-González, J. Solid-State Fermentation: Physiology of Solid Medium, Its Molecular Basis and Applications. Process Biochem. 2012, 47, 175–185.
- Chaudhary, V.; Katyal, P.; Puniya, A.K.; Panwar, H.; Arora, M.; Kaur, J.; Rokana, N.; Wakchaure, N.; Raposo, A.; Raheem, D.; et al. Pilot-Scale Process to Produce Bio-Pigment from Monascus Purpureus Using Broken Rice as Substrate for Solid-State Fermentation. Eur. Food Res. Technol. 2023, 249, 1845–1855.
- Srianta, I.; Zubaidah, E.; Estiasih, T.; Yamada, M.; Harijono. Comparison of Monascus purpureus Growth, Pigment Production and Composition on Different Cereal Substrates with Solid State Fermentation. Biocatal. Agric. Biotechnol. 2016, 7, 181–186.
- Domínguez, M.; Mejía, A.; Revah, S.; Barrios-González, J. Optimization of Bagasse, Nutrients and Initial Moisture Ratios on the Yield of Penicillin in Solid-State Fermentation. World J. Microbiol. Biotechnol. 2001, 17, 751–756.
- Domínguez, M.; Mejía, A.; Barrios-González, J. Respiration Studies of Penicillin Solid-State Fermentation. J. Biosci. Bioeng. 2000, 89, 409–413.
- López-Calleja, A.C.; Cuadra, T.; Barrios-González, J.; Fierro, F.; Fernández, F.J. Solid-State and Submerged Fermentations Show Different Gene Expression Profiles in Cephalosporin C Production by Acremonium chrysogenum. Microb. Physiol. 2012, 22, 126–134.
- Cuadra, T.; Fernández, F.J.; Tomasini, A.; Barrios-González, J. Influence of PH Regulation and Nutrient Content on Cephalosporin C Production in Solid-State Fermentation by Acremonium Chrysogenum C10. Lett. Appl. Microbiol. 2007, 46, 216–220.
- El-Housseiny, G.S.; Ibrahim, A.A.; Yassien, M.A.; Aboshanab, K.M. Production and Statistical Optimization of Paromomycin by Streptomyces rimosus NRRL 2455 in Solid State Fermentation. BMC Microbiol. 2021, 21, 34.
- Machado, I.; Teixeira, J.A.; Rodríguez-Couto, S. Semi-Solid-State Fermentation: A Promising Alternative for Neomycin Production by the Actinomycete Streptomyces fradiae. J. Biotechnol. 2013, 165, 195–200.
- Vastrad, B.M.; Neelagund, S.E.; Iiger, S.R.; Godbole, A.M.; Kulkarni, V. Improved Rifamycin B Production by Nocardia mediterranei MTCC 14 under Solid-State Fermentation through Process Optimization. Biochem. Res. Int. 2014, 2014, 1–13.
- Lucas, E.M.; Machado, Y.; Ferreira, A.A.; Dolabella, L.M.; Takahashi, J.A. Improved Production of Pharmacologically-Active Sclerotiorin by Penicillium Sclerotiorum. 2010, Volume 9. Available online: http://www.tjpr.org (accessed on 15 June 2023).
- Saykhedkar, S.S.; Singhal, R.S. Solid-State Fermentation for Production of Griseofulvin on Rice Bran Using Penicillium griseofulvum. Biotechnol. Prog. 2004, 20, 1280–1284.
- Zeng, X.; Miao, W.; Zeng, H.; Zhao, K.; Zhou, Y.; Zhang, J.; Zhao, Q.; Tursun, D.; Xu, D.; Li, F. Production of Natamycin by Streptomyces gilvosporeus Z28 through Solid-State Fermentation Using Agro-Industrial Residues. Bioresour. Technol. 2019, 273, 377–385.
- Baños, J.G.; Tomasini, A.; Szakács, G.; Barrios-González, J. High Lovastatin Production by Aspergillus terreus in Solid-State Fermentation on Polyurethane Foam: An Artificial Inert Support. J. Biosci. Bioeng. 2009, 108, 105–110.
- Ávila, N.; Tarragó-Castellanos, M.R.; Barrios-González, J. Environmental Cues That Induce the Physiology of Solid Medium: A Study on Lovastatin Production by Aspergillus terreus. J. Appl. Microbiol. 2017, 122, 1029–1038.
- Pérez-Sánchez, A.; Uribe-Carvajal, S.; Cabrera-Orefice, A.; Barrios-González, J. Key Role of Alternative Oxidase in Lovastatin Solid-State Fermentation. Appl. Microbiol. Biotechnol. 2017, 101, 7347–7356.
- Shaligram, N.S.; Singh, S.K.; Singhal, R.S.; Pandey, A.; Szakacs, G. Compactin Production Studies Using Penicillium brevicompactum Under Solid-State Fermentation Conditions. Appl. Biochem. Biotechnol. 2009, 159, 505–520.
- Zhang, B.-B.; Xing, H.-B.; Jiang, B.-J.; Chen, L.; Xu, G.-R.; Jiang, Y.; Zhang, D.-Y. Using Millet as Substrate for Efficient Production of Monacolin K by Solid-State Fermentation of Monascus ruber. J. Biosci. Bioeng. 2018, 125, 333–338.
- Liu, X.; Sun, A.; Li, Q.; Du, Y.; Zhao, T. A Systematic Study of the Production of Monacolin K by Solid State Fermentation of Monascus ruber. AMB Express 2022, 12, 29.
- Zouari, R.; Ellouze-Chaabouni, S.; Ghribi-Aydi, D. Optimization of Bacillus subtilis SPB1 Biosurfactant Production Under Solid-State Fermentation Using By-Products of a Traditional Olive Mill Factory. Achiev. Life Sci. 2014, 8, 162–169.
- Zhu, Z.; Zhang, F.; Wei, Z.; Ran, W.; Shen, Q. The Usage of Rice Straw as a Major Substrate for the Production of Surfactin by Bacillus amyloliquefaciens XZ-173 in Solid-State Fermentation. J. Env. Environ. Manag. 2013, 127, 96–102.
- Piedrahíta-Aguirre, C.A.; Bastos, R.G.; Carvalho, A.L.; Monte Alegre, R. The Influence of Process Parameters in Production of Lipopeptide Iturin A Using Aerated Packed Bed Bioreactors in Solid-State Fermentation. Bioprocess. Biosyst. Eng. 2014, 37, 1569–1576.
- Gong, Z.; He, Q.; Che, C.; Liu, J.; Yang, G. Optimization and Scale-up of the Production of Rhamnolipid by Pseudomonas aeruginosa in Solid-State Fermentation Using High-Density Polyurethane Foam as an Inert Support. Bioprocess. Biosyst. Eng. 2020, 43, 385–392.
- Dabaghi, S.; Ataei, S.A.; Taheri, A. Production of Rhamnolipid Biosurfactants in Solid-State Fermentation: Process Optimization and Characterization Studies. BMC Biotechnol. 2023, 23, 2.
- Jiménez-Peñalver, P.; Castillejos, M.; Koh, A.; Gross, R.; Sánchez, A.; Font, X.; Gea, T. Production and Characterization of Sophorolipids from Stearic Acid by Solid-State Fermentation, a Cleaner Alternative to Chemical Surfactants. J. Clean. Prod. 2018, 172, 2735–2747.
- Rodríguez, A.; Gea, T.; Font, X. Sophorolipids Production from Oil Cake by Solid-State Fermentation. Inventory for Economic and Environmental Assessment. Front. Chem. Eng. 2021, 3, 2752.
- Mehmood, T.; Saleem, F.; Javed, S.; Nawaz, S.; Sultan, A.; Safdar, A.; Ullah, A.; Waseem, R.; Saeed, S.; Abbas, M.; et al. Biotransformation of Agricultural By-Products into Biovanillin through Solid-State Fermentation (SSF) and Optimization of Different Parameters Using Response Surface Methodology (RSM). Fermentation 2022, 8, 206.
- Mehmood, T.; Ahmed, S.; Waseem, R.; Saeed, S.; Ahmed, W.; Irfan, M.; Ullah, A. Valorization of Fruit Peels into Biovanillin and Statistical Optimization of Process Using Enterobacter hormaechei through Solid-State Fermentation. Fermentation 2022, 8, 40.
- Mehmood, T.; Saeed, S.; Hussain, N.; Waseem, R. Biotransformation of Wheat Straw into Biovanillin by Solid-State Fermentation and Optimization of Conditions Parameters through Response Surface Methodology. Biomass Convers. Biorefin 2022, 19, 10.
- Saeed, S.; Aslam, S.; Mehmood, T.; Naseer, R.; Nawaz, S.; Mujahid, H.; Firyal, S.; Anjum, A.A.; Sultan, A. Production of Gallic Acid Under Solid-State Fermentation by Utilizing Waste from Food Processing Industries. Waste Biomass Valorization 2021, 12, 155–163.
- Wu, C.-Y.; Liang, C.-H.; Ou, C.-H.; Liang, Z.-C. Zinc Ion Addition to Grain Media Enhanced Hispidin Production during Solid-State Fermentation of Phellinus linteus. J. Taiwan. Inst. Chem. Eng. 2021, 121, 101–107.
- Patel, G.; Patil, M.D.; Soni, S.; Chisti, Y.; Banerjee, U.C. Production of Mycophenolic Acid by Penicillium brevicompactum Using Solid State Fermentation. Appl. Biochem. Biotechnol. 2017, 182, 97–109.
- Survase, S.A. A Novel Medium for the Enhanced Production of Cyclosporin A ByTolypocladium inflatum MTCC 557 Using Solid State Fermentation. J. Microbiol. Biotechnol. 2009, 19, 462–467.
- Gelmi, C.; Pérez-Correa, R.; Agosin, E. Modelling Gibberella fujikuroi Growth and GA3 Production in Solid-State Fermentation. Process Biochem. 2002, 37, 1033–1040.
- Doi, Y.; Wakana, D.; Takeda, H.; Tanaka, E.; Hosoe, T. Production of Ergot Alkaloids by the Japanese Isolate Claviceps purpurea Var. Agropyri on Rice Medium. Adv. Microbiol. 2022, 12, 254–269.
- Dufossé, L.; Galaup, P.; Yaron, A.; Arad, S.M.; Blanc, P.; Chidambara Murthy, K.N.; Ravishankar, G.A. Microorganisms and Microalgae as Sources of Pigments for Food Use: A Scientific Oddity or an Industrial Reality? Trends Food Sci. Technol. 2005, 16, 389–406.
- Arumugam, G.K.; Selvaraj, V.; Gopal, D.; Ramalingam, K. Solid-State Fermentation of Agricultural Residues for the Production of Antibiotics. In Biotransformation of Waste Biomass into High Value Biochemicals; Springer: New York, NY, USA, 2014; pp. 139–162.
- Barrios-González, J.; Pérez-Sánchez, A.; Bibián, M.E. New Knowledge about the Biosynthesis of Lovastatin and Its Production by Fermentation of Aspergillus terreus. Appl. Microbiol. Biotechnol. 2020, 104, 8979–8998.
- Suryanarayan, S. Current Industrial Practice in Solid State Fermentations for Secondary Metabolite Production: The Biocon India Experience. Biochem. Eng. J. 2003, 13, 189–195.
- Banat, I.M.; Carboué, Q.; Saucedo-Castañeda, G.; de Jesús Cázares-Marinero, J. Biosurfactants: The Green Generation of Speciality Chemicals and Potential Production Using Solid-State Fermentation (SSF) Technology. Bioresour. Technol. 2021, 320, 124222.
- Pascoalino, L.A.; Pires, T.C.S.P.; Taofiq, O.; Ferreira, I.C.F.R.; Barros, L.; Reis, F.S. Biochemistry of Secondary Metabolism of Fungi. In Natural Secondary Metabolites; Springer International Publishing: Cham, Switzerland, 2023; pp. 437–474.
- Saltveit, M.E. Synthesis and Metabolism of Phenolic Compounds. In Fruit and Vegetable Phytochemicals; John Wiley & Sons, Ltd.: Chichester, UK, 2017; pp. 115–124.
- Zurbriggen, A.; Kirst, H.; Melis, A. Isoprene Production Via the Mevalonic Acid Pathway in Escherichia coli (Bacteria). Bioenergy Res. 2012, 5, 814–828.
- Francenia Santos-Sánchez, N.; Salas-Coronado, R.; Hernández-Carlos, B.; Villanueva-Cañongo, C. Shikimic Acid Pathway in Biosynthesis of Phenolic Compounds. In Plant Physiological Aspects of Phenolic Compounds; IntechOpen: Rijeka, Croatia, 2019.
- Sheng, Q.; Yi, L.; Zhong, B.; Wu, X.; Liu, L.; Zhang, B. Shikimic Acid Biosynthesis in Microorganisms: Current Status and Future Direction. Biotechnol. Adv. 2023, 62, 108073.
- Emiliani, G.; Fondi, M.; Fani, R.; Gribaldo, S. A Horizontal Gene Transfer at the Origin of Phenylpropanoid Metabolism: A Key Adaptation of Plants to Land. Biol. Direct 2009, 4, 7.
- Wu, S.; Chen, W.; Lu, S.; Zhang, H.; Yin, L. Metabolic Engineering of Shikimic Acid Biosynthesis Pathway for the Production of Shikimic Acid and Its Branched Products in Microorganisms: Advances and Prospects. Molecules 2022, 27, 4779.
- Ghosh, S.; Chisti, Y.; Banerjee, U.C. Production of Shikimic Acid. Biotechnol. Adv. 2012, 30, 1425–1431.
- Dhaniaputri, R.; Suwono, H.; Amin, M.; Lukiati, B. Introduction to Plant Metabolism, Secondary Metabolites Biosynthetic Pathway, and In-Silico Molecular Docking for Determination of Plant Medicinal Compounds: An Overview. In Proceedings of the 7th International Conference on Biological Science (ICBS 2021), Online, 14-15 October 2021.
- Matulja, D.; Vranješević, F.; Kolympadi Markovic, M.; Pavelić, S.K.; Marković, D. Anticancer Activities of Marine-Derived Phenolic Compounds and Their Derivatives. Molecules 2022, 27, 1449.
- Diner, B.A.; Fan, J.; Scotcher, M.C.; Wells, D.H.; Whited, G.M. Synthesis of Heterologous Mevalonic Acid Pathway Enzymes in Clostridium ljungdahlii for the Conversion of Fructose and of Syngas to Mevalonate and Isoprene. Appl. Env. Environ. Microbiol. 2018, 84, e01723-17.
- Boronat, A.; Rodríguez-Concepción, M. Terpenoid Biosynthesis in Prokaryotes. In Biotechnology of Isoprenoids; Springer: Cham, Switzerland, 2014; pp. 3–18.
- Karlic, H.; Varga, F. Mevalonate Pathway. In Encyclopedia of Cancer; Boffetta, P., Hainaut, P., Eds.; Academic Press: Oxford, UK, 2019; pp. 445–447.
- Zeng, L.; Dehesh, K. The Eukaryotic MEP-Pathway Genes Are Evolutionarily Conserved and Originated from Chlaymidia and Cyanobacteria. BMC Genom. 2021, 22, 137.
- Zhao, L.; Chang, W.; Xiao, Y.; Liu, H.; Liu, P. Methylerythritol Phosphate Pathway of Isoprenoid Biosynthesis. Annu. Rev. Biochem. 2013, 82, 497–530.
- Bode, H.B.; Müller, R. Possibility of Bacterial Recruitment of Plant Genes Associated with the Biosynthesis of Secondary Metabolites. Plant Physiol. 2003, 132, 1153–1161.
- Hu, M.; Chen, S.; Ni, Y.; Wei, W.; Mao, W.; Ge, M.; Qian, X. CRISPR/Cas9-Mediated Genome Editing in Vancomycin-Producing Strain Amycolatopsis keratiniphila. Front. Bioeng. Biotechnol. 2023, 11, 1141176.
- Qian, H.; Wei, W.; Chen, X.-A.; Mo, X.-T.; Ge, M.; Zhao, Q.-W.; Li, Y.-Q. Strategy for Producing the High-Quality Glycopeptide Antibiotic A82846B in Amycolatopsis Orientalis Based on the CRISPR-Cas12a System. ACS Synth. Biol. 2021, 10, 3009–3016.
- Vásquez-Bonilla, J.N.; Barranco-Florido, J.E.; Ponce-Alquicira, E.; Rincón-Guevara, M.A.; Loera, O. Improvement of Beauvericin Production by Fusarium Oxysporum AB2 under Solid-State Fermentation Using an Optimised Liquid Medium and Co-Cultures. Mycotoxin Res. 2022, 38, 175–183.
This entry is offline, you can click here to edit this entry!
