The sudden outbreak of the COVID-19 pandemic led to a huge concern globally because of the astounding increase in mortality rates worldwide. The medical imaging computed tomography technique, whole-genome sequencing, and electron microscopy are the methods generally used for the screening and identification of the SARS-CoV-2 virus.
- COVID-19 virus
- optical spectroscopy
- optical biosensors
- optical diagnosis
- optical therapy
- artificial intelligence
- Internet of Things
- machine learning
1. Introduction
In recent times, COVID-19 played havoc, causing escalating mortality rates globally. According to the World Health Organization (WHO), on 21 February 2023, 757,264,511 confirmed cases and 6,850,594 deaths related to COVID-19 were reported globally [1]. The history of viral pandemic diseases, including the Spanish flu, which originated in 1918, severe acute respiratory syndrome (SARS), and the Middle East respiratory syndrome (MERS), which originated a decade ago, and the recent emergence of a novel coronavirus caused by the SARS-CoV-2 virus.
The MERS, SARS, and SARS-CoV-2 viruses are considered the most lethal respiratory diseases transmitted by zoonotic transmission, leading to death in patients with severe comorbid conditions [2][3]. They are highly contagious among humans. Each type of virus is distinctively characterized based on its surface proteins and lipid profiles. Coronavirus belongs to the subfamily Coronaviridae and is an enveloped RNA virus 100–160 nm in diameter, with a spherical structure. This subfamily, based on phylogenetic relationships and genomic structures, is classified into four groups: (i) alpha coronavirus, (ii) beta coronavirus, which infects mammals, (iii) gamma coronavirus, which infects aves, and (iv) delta coronavirus, which infects both aves and mammals [2]. Coronavirus has the single-stranded positive-sense RNA (ssRNA) genetic material and the largest genome of 26.4–31.7 kb, which encodes the structural proteins such as spike glycoprotein (S), membrane glycoprotein (M), nucleocapsid interrupts phosphor protein (N), and envelope (E) protein [2][3][4]. The genome of SARS-CoV-2 shares 82% of its sequence identity with SARS-CoV and MERS-CoV and more than 90% of its sequence identity with that of structural proteins and essential enzymes. Coronavirus uses the spike protein (S) to bind to the receptor of the host cell surface and initiate infection. The mechanism of host entry differs for different coronaviruses [5]. A more detailed discussion on the genomics, proteomics, and mechanism of pathogenesis of SARS-CoV-2 has been reported as a breakthrough therapy [2].
2. Optical Biosensors
2.1. Spectroscopy and Nanomaterials-Based Optical Biosensors
2.1.1. Raman Spectroscopy
2.1.2. Near-Infrared and Fourier Transform Infrared Spectroscopy
2.1.3. Gold-Nanoparticle-Based Surface Plasmon Resonance
2.1.4. Gold-Nanoparticle-Based Surface-Enhanced Raman Spectroscopy
2.1.5. Magnetic-Nanoparticle-Based Fluorescence Biosensors
2.2. Interferometry-Based Optical Biosensors
2.2.1. Photonic Crystal Biosensors
2.2.2. Back Focal Plane Interferometry
2.2.3. Mach–Zehnder Interferometry
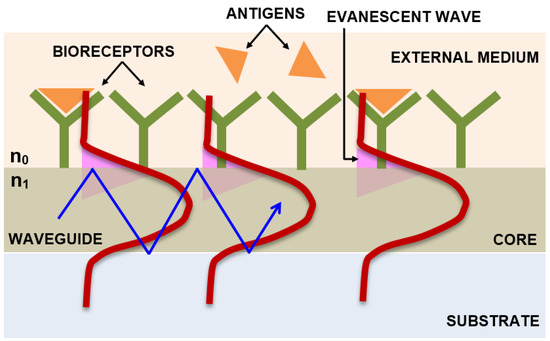
2.2.4. Integrated Optical Young’s Interferometry
2.2.5. Interferometry Reflectance Imaging Sensor
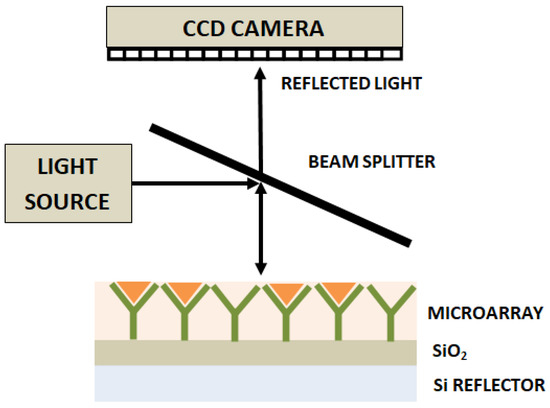
2.2.6. Hartman Interferometer
2.2.7. Liquid Core Optical Ring Resonator
2.3. Lab-on-a-Chip-Based Optical Biosensors
2.4. Smartphone-Based Portable Optical Biosensors
2.5. Artificial-Intelligence-Based Smart Optical Biosensors
3. Conclusions
This entry is adapted from the peer-reviewed paper 10.3390/diagnostics13142418
References
- WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 16 April 2023).
- Naqvi, A.A.T.; Fatima, K.; Mohammad, T.; Fatima, U.; Singh, I.K.; Singh, A.; Atif, S.M.; Hariprasad, G.; Hasan, G.M.; Hassan, M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta BBA-Mol. Basis Dis. 2002, 1866, 165878.
- Cui, J.; Li, F.; Shi, Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192.
- Pellett, P.E.; Mitra, S.; Holland, T.C. Basics of virology. Handb. Clin. Neurol. 2014, 123, 45–66.
- Lau, Y.L.; Peiris, J.M. Pathogenesis of severe acute respiratory syndrome. Curr. Opin. Immunol. 2005, 17, 404–410.
- Santos, M.C.; Morais, C.L.; Nascimento, Y.M.; Araujo, J.M.; Lima, K.M. Spectroscopy with computational analysis in virological studies: A decade (2006–2016). TrAC Trends Anal. Chem. 2017, 97, 244–256.
- Chaudhary, I.; Jackson, N.; Denning, D.; O’Neill, L.; Byrne, H.J. Contributions of vibrational spectroscopy to virology: A review. Clin. Spectrosc. 2022, 4, 100022.
- Moor, K.; Ohtani, K.; Myrzakozha, D.; Zhanserkenova, O.; Andriana, B.B.; Sato, H. Noninvasive and label-free determination of virus infected cells by Raman spectroscopy. J. Biomed. Opt. 2014, 19, 067003.
- Santos, M.C.; Monteiro, J.D.; Araújo, J.M.; Lima, K.M. Molecular fluorescence spectroscopy with multi-way analysis techniques detects spectral variations distinguishing uninfected serum versus dengue or chikungunya viral infected samples. Sci. Rep. 2020, 10, 13758.
- Rodríguez-Casado, A.; Bartolomé, J.; Carreño, V.; Molina, M.; Carmona, P. Structural characterization of the 5′ untranslated RNA of hepatitis C virus by vibrational spectroscopy. Biophys. Chem. 2006, 124, 73–79.
- Kim, H.; Hwang, J.; Kim, J.H.; Lee, S.; Kang, M. Sensitive detection of multiple fluoresence probes based on surface-enhanced Raman scattering (sers) for mers-cov. In Proceedings of the 2019 IEEE 14th International Conference on Nano/Micro Engineered and 776 Molecular Systems (NEMS), Bangkok, Thailand, 11–14 April 2019; pp. 498–501.
- Sakudo, A.; Suganuma, Y.; Sakima, R.; Ikuta, K. Diagnosis of HIV-1 infection by near-infrared spectroscopy: Analysis using molecular clones of various HIV-1 subtypes. Clin. Chim. Acta 2012, 413, 467–472.
- Amathieu, R.; Triba, M.N.; Goossens, C.; Bouchemal, N.; Nahon, P.; Savarin, P.; Le Moyec, L. Nuclear magnetic resonance based metabolomics and liver diseases: Recent advances and future clinical applications. World J. Gastroenterol. 2016, 22, 417.
- Slupsky, C.M. Nuclear magnetic resonance-based analysis of urine for the rapid etiological diagnosis of pneumonia. Expert Opin. Med. Diagn. 2011, 5, 63–73.
- Lambert, P.J.; Whitman, A.G.; Dyson, O.F.; Akula, S.M. Raman spectroscopy: The gateway into tomorrow’s virology. Virol. J. 2006, 3, 51.
- Salman, A.; Shufan, E.; Zeiri, L.; Huleihel, M. Characterization and detection of Vero cells infected with Herpes Simplex Virus type 1 using Raman spectroscopy and advanced statistical methods. Methods 2014, 68, 364–370.
- Kelly, J.G.; Trevisan, J.; Scott, A.D.; Carmichael, P.L.; Pollock, H.M.; Martin-Hirsch, P.L.; Martin, F.L. Biospectroscopy to metabolically profile biomolecular structure: A multistage approach linking computational analysis with biomarkers. J. Proteome Res. 2011, 10, 1437–1448.
- Saade, J.; Pacheco, M.T.T.; Rodrigues, M.R.; Silveira, L., Jr. Identification of hepatitis C in human blood serum by near-infrared Raman spectroscopy. Spectroscopy 2008, 22, 387–395.
- Howes, P.D.; Chandrawati, R.; Stevens, M.M. Colloidal nanoparticles as advanced biological sensors. Science 2014, 346, 1247390.
- Baptista, P.; Pereira, E.; Eaton, P.; Doria, G.; Miranda, A.; Gomes, I.; Quaresma, P.; Franco, R. Gold nanoparticles for the development of clinical diagnosis methods. Anal. Bioanal. Chem. 2008, 391, 943–950.
- Neng, J.; Harpster, M.H.; Wilson, W.C.; Johnson, P.A. Surface-enhanced Raman scattering (SERS) detection of multiple viral antigens using magnetic capture of SERS-active nanoparticles. Biosens. Bioelectron. 2013, 41, 316–321.
- Cao, Y.C.; Jin, R.; Mirkin, C.A. Nanoparticles with Raman spectroscopic fingerprints for DNA and RNA detection. Science 2002, 297, 1536–1540.
- Griffin, J.; Singh, A.K.; Senapati, D.; Rhodes, P.; Mitchell, K.; Robinson, B.; Yu, E.; Ray, P.C. Size-and distance-dependent nanoparticle surface-energy transfer (NSET) method for selective sensing of hepatitis C virus RNA. Chem. A Eur. J. 2009, 15, 342–351.
- Lu, X.; Dong, X.; Zhang, K.; Han, X.; Fang, X.; Zhang, Y. A gold nanorods-based fluorescent biosensor for the detection of hepatitis B virus DNA based on fluorescence resonance energy transfer. Analyst 2013, 138, 642–650.
- Nasrin, F.; Chowdhury, A.D.; Takemura, K.; Lee, J.; Adegoke, O.; Deo, V.K.; Abe, F.; Suzuki, T.; Park, E.Y. Single-step detection of norovirus tuning localized surface plasmon resonance-induced optical signal between gold nanoparticles and quantum dots. Biosens. Bioelectron. 2018, 122, 16–24.
- Chang, Y.F.; Wang, S.F.; Huang, J.C.; Su, L.C.; Yao, L.; Li, Y.C.; Wu, S.C.; Chen, Y.M.A.; Hsieh, J.P.; Chou, C. Detection of swine-origin influenza A (H1N1) viruses using a localized surface plasmon coupled fluorescence fiber-optic biosensor. Biosens. Bioelectron. 2010, 26, 1068–1073.
- Ganbold, E.O.; Kang, T.; Lee, K.; Lee, S.Y.; Joo, S.W. Aggregation effects of gold nanoparticles for single-base mismatch detection in influenza A (H1N1) DNA sequences using fluorescence and Raman measurements. Colloids Surf. B Biointerfaces 2012, 93, 148–153.
- Draz, M.S.; Fang, B.A.; Li, L.; Chen, Z.; Wang, Y.; Xu, Y.; Yang, J.; Killeen, K.; Chen, F.F. Hybrid nanocluster plasmonic resonator for immunological detection of hepatitis B virus. ACS Nano 2012, 6, 7634–7643.
- Nasrin, F.; Chowdhury, A.D.; Takemura, K.; Kozaki, I.; Honda, H.; Adegoke, O.; Park, E.Y. Fluorometric virus detection platform using quantum dots-gold nanocomposites optimizing the linker length variation. Anal. Chim. Acta 2020, 1109, 148–157.
- Li, Y.; Lu, C.; Zhou, S.; Fauconnier, M.L.; Gao, F.; Fan, B.; Lin, J.; Wang, F.; Zheng, J. Sensitive and simultaneous detection of different pathogens by surface-enhanced Raman scattering based on aptamer and Raman reporter co-mediated gold tags. Sens. Actuators B Chem. 2020, 317, 128182.
- Ngo, H.T.; Wang, H.-N.; Fales, A.M.; Nicholson, B.P.; Woods, C.W.; VoDinh, T. DNA bioassay-on-chip using SERS detection for dengue diagnosis. Analyst 2014, 139, 5655–5659.
- Maddali, H.; Miles, C.E.; Kohn, J.; O’Carroll, D.M. Optical biosensors for virus detection: Prospects for SARS-CoV-2/COVID-19. ChemBioChem 2021, 22, 1176–1189.
- Ahmed, S.R.; Kang, S.W.; Oh, S.; Lee, J.; Neethirajan, S. Chiral zirconium quantum dots: A new class of nanocrystals for optical detection of coronavirus. Heliyon 2018, 4, e00766.
- Xie, B.P.; Qiu, G.H.; Hu, P.P.; Liang, Z.; Liang, Y.M.; Sun, B.; Bai, L.P.; Jiang, Z.H.; Chen, J.X. Simultaneous detection of Dengue and Zika virus RNA sequences with a three-dimensional Cu-based zwitterionic metal–organic framework, comparison of single and synchronous fluorescence analysis. Sens. Actuators B Chem. 2018, 254, 1133–1140.
- Fan, X.; White, I.M.; Shopova, S.I.; Zhu, H.; Suter, J.D.; Sun, Y. Sensitive optical biosensors for unlabeled targets: A review. Anal. Chim. Acta 2008, 620, 8–26.
- John, P.; Vasa, N.J.; Unni, S.N.; Rao, S.R. Glucose sensing in oral mucosa simulating phantom using differential absorption based frequency domain low-coherence interferometry. Appl. Opt. 2017, 56, 8257–8265.
- John, P.; Vasa, N.J.; Sujatha, N. Glucose sensing in the anterior chamber of the human eye model using supercontinuum source based dual wavelength low coherence interferometry. Sens. Bio-Sens. Res. 2019, 23, 100277.
- Goddard, J.M.; Mandal, S.; Nugen, S.R.; Baeumner, A.J.; Erickson, D. Biopatterning for label-free detection. Colloids Surf. B Biointerfaces 2010, 76, 375–380.
- Huang, M.C.; Mateus, C.F.; Foley, J.E.; Beatty, R.; Cunningham, B.T.; Chang-Hasnain, C.J. VCSEL optoelectronic biosensor for detection of infectious diseases. IEEE Photonics Technol. Lett. 2008, 20, 443–445.
- Pang, Y.; Song, H.; Cheng, W. Using optical trap to measure the refractive index of a single animal virus in culture fluid with high precision. Biomed. Opt. Express 2016, 7, 1672–1689.
- Xu, J.; Suarez, D.; Gottfried, D.S. Detection of avian influenza virus using an interferometric biosensor. Anal. Bioanal. Chem. 2007, 389, 1193–1199.
- Ymeti, A.; Greve, J.; Lambeck, P.V.; Wink, T.; van Hövell, S.W.; Beumer, T.A.; Wijn, R.R.; Heideman, R.G.; Subramaniam, V.; Kanger, J.S. Fast, ultrasensitive virus detection using a young interferometer sensor. Nano Lett. 2007, 7, 394–397.
- Ymeti, A.; Subramaniam, V.; Beumer, T.A.; Kanger, J.S. An ultrasensitive young interferometer handheld sensor for rapid virus detection. Expert Rev. Med. Devices 2007, 4, 447–454.
- Avci, O.; Lortlar, Ü.N.; Yalçın Özkumur, A.; Ünlü, M.S. Interferometric reflectance imaging sensor (IRIS)—A platform technology for multiplexed diagnostics and digital detection. Sensors 2015, 15, 17649–17665.
- Schneider, B.H.; Edwards, J.G.; Hartman, N.F. Hartman interferometer: Versatile integrated optic sensor for label-free, real-time quantification of nucleic acids, proteins, and pathogens. Clin. Chem. 1997, 43, 1757–1763.
- Fan, X.; White, I.M.; Zhu, H.; Suter, J.D.; Oveys, H. Overview of novel integrated optical ring resonator bio/chemical sensors. In Laser Resonators and Beam Control IX; SPIE: Bellingham, Washington USA, 2007; Volume 6452, pp. 166–185.
- White, I.M.; Oveys, H.; Fan, X.; Smith, T.L.; Zhang, J. Integrated multiplexed biosensors based on liquid core optical ring resonators and antiresonant reflecting optical waveguides. Appl. Phys. Lett. 2006, 89, 191106.
- Zhu, H.; White, I.M.; Suter, J.D.; Zourob, M.; Fan, X. Opto-fluidic micro-ring resonator for sensitive label-free viral detection. Analyst 2008, 133, 356–360.
- Pires, N.M.M.; Dong, T.; Hanke, U.; Hoivik, N. Recent developments in optical detection technologies in lab-on-a-chip devices for biosensing applications. Sensors 2014, 14, 15458–15479.
- Zhu, H.; Fohlerová, Z.; Pekárek, J.; Basova, E.; Neužil, P. Recent advances in lab-on-a-chip technologies for viral diagnosis. Biosens. Bioelectron. 2020, 153, 112041.
- Ahrberg, C.D.; Manz, A.; Neuzil, P. Palm-sized device for point-of-care Ebola detection. Anal. Chem. 2016, 88, 4803–4807.
- Li, Z.; Zhao, J.; Wu, X.; Zhu, C.; Liu, Y.; Wang, A.; Deng, G.; Zhu, L. A rapid microfluidic platform with real-time fluorescence detection system for molecular diagnosis. Biotechnol. Biotechnol. Equip. 2019, 33, 223–230.
- Bedin, F.; Boulet, L.; Voilin, E.; Theillet, G.; Rubens, A.; Rozand, C. Paper-based point-of-care testing for cost-effective diagnosis of acute flavivirus infections. J. Med. Virol. 2017, 89, 1520–1527.
- Dincer, C.; Bruch, R.; Costa-Rama, E.; Fernández-Abedul, M.T.; Merkoçi, A.; Manz, A.; Urban, G.A.; Güder, F. Disposable sensors in diagnostics, food, and environmental monitoring. Adv. Mater. 2019, 31, 1806739.
- Quesada-González, D.; Merkoçi, A. Nanomaterial-based devices for point-of-care diagnostic applications. Chem. Soc. Rev. 2018, 47, 4697–4709.
- Cho, S.; Islas-Robles, A.; Nicolini, A.M.; Monks, T.J.; Yoon, J.Y. In situ, dual-mode monitoring of organ-on-a-chip with smartphone-based fluorescence microscope. Biosens. Bioelectron. 2016, 86, 697–705.
- Roblyer, D. Perspective on the increasing role of optical wearables and remote patient monitoring in the COVID-19 era and beyond. J. Biomed. Opt. 2020, 25, 102703.
- El Asnaoui, K.; Chawki, Y. Using X-ray images and deep learning for automated detection of coronavirus disease. J. Biomol. Struct. Dyn. 2021, 39, 3615–3626.
- Hemdan, E.E.D.; Shouman, M.A.; Karar, M.E. Covidx-net: A framework of deep learning classifiers to diagnose COVID-19 in X-ray images. arXiv 2020, arXiv:2003.11055.
- Shi, F.; Wang, J.; Shi, J.; Wu, Z.; Wang, Q.; Tang, Z.; He, K.; Shi, Y.; Shen, D. Review of artificial intelligence techniques in imaging data acquisition, segmentation, and diagnosis for COVID-19. IEEE Rev. Biomed. Eng. 2020, 14, 4–15.
- Xu, X.; Jiang, X.; Ma, C.; Du, P.; Li, X.; Lv, S.; Yu, L.; Ni, Q.; Chen, Y.; Su, J.; et al. A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering 2020, 6, 1122–1129.
- Jamshidi, M.; Lalbakhsh, A.; Talla, J.; Peroutka, Z.; Hadjilooei, F.; Lalbakhsh, P.; Jamshidi, M.; La Spada, L.; Mirmozafari, M.; Dehghani, M.; et al. Artificial intelligence and COVID-19: Deep learning approaches for diagnosis and treatment. IEEE Access 2020, 8, 109581–109595.
- O’Connor, T.; Javidi, B. COVID-19 screening with digital holographic microscopy using intra-patient probability functions of spatio-temporal bio-optical attributes. Biomed. Opt. Express 2022, 13, 5377–5389.
- Shamout, F.E.; Shen, Y.; Wu, N.; Kaku, A.; Park, J.; Makino, T.; Jastrzębski, S.; Witowski, J.; Wang, D.; Zhang, B.; et al. An artificial intelligence system for predicting the deterioration of COVID-19 patients in the emergency department. NPJ Digit. Med. 2021, 4, 80.
- Barnawi, A.; Chhikara, P.; Tekchandani, R.; Kumar, N.; Alzahrani, B. Artificial intelligence-enabled Internet of Things-based system for COVID-19 screening using aerial thermal imaging. Future Gener. Comput. Syst. 2021, 124, 119–132.
- Carvalho, L.F.d.C.e.S.; Nogueira, M.S. Optical techniques for fast screening-Towards prevention of the coronavirus COVID-19 outbreak. Photodiagnosis Photodyn. Ther. 2020, 30, 101765.
- Rai, M.; Deshmukh, S.D.; Ingle, A.P.; Gupta, I.R.; Galdiero, M.; Galdiero, S. Metal nanoparticles: The protective nanoshield against virus infection. Crit. Rev. Microbiol. 2016, 42, 46–56.
- Łoczechin, A.; Séron, K.; Barras, A.; Giovanelli, E.; Belouzard, S.; Chen, Y.T.; Metzler-Nolte, N.; Boukherroub, R.; Dubuisson, J.; Szunerits, S. Functional carbon quantum dots as medical countermeasures to human coronavirus. ACS Appl. Mater. Interfaces 2019, 11, 42964–42974.
- Wainwright, M. Local treatment of viral disease using photodynamic therapy. Int. J. Antimicrob. Agents 2003, 21, 510–520.
- Kharkwal, G.B.; Sharma, S.K.; Huang, Y.Y.; Dai, T.; Hamblin, M.R. Photodynamic therapy for infections: Clinical applications. Lasers Surg. Med. 2011, 43, 755–767.
- Agostinis, P.; Berg, K.; Cengel, K.A.; Foster, T.H.; Girotti, A.W.; Gollnick, S.O.; Hahn, S.M.; Hamblin, M.R.; Juzeniene, A.; Kessel, D.; et al. Photodynamic therapy of cancer: An update. CA Cancer J. Clin. 2011, 61, 250–281.
- Fekrazad, R. Photobiomodulation and antiviral photodynamic therapy as a possible novel approach in COVID-19 management. Photobiomodulation Photomed. Laser Surg. 2020, 38, 255–257.
