During early mammalian embryonic development, fertilized one-cell embryos develop into pre-implantation blastocysts and subsequently establish three germ layers through gastrulation during post-implantation development. Stem cells have emerged as a powerful tool to study embryogenesis and gastrulation without the need for eggs, allowing for the generation of embryo-like structures known as synthetic embryos or embryoids. These in vitro models closely resemble early embryos in terms of morphology and gene expression and provide a faithful recapitulation of early pre- and post-implantation embryonic development. Synthetic embryos can be generated through a combinatorial culture of three blastocyst-derived stem cell types, such as embryonic stem cells, trophoblast stem cells, and extraembryonic endoderm cells, or totipotent-like stem cells alone.
- synthetic embryo
- embryogenesis
- gastrulation
- stem cells
1. Introduction
2. Early Embryonic Development in Mice
2.1. Pre-Implantation Embryonic Development
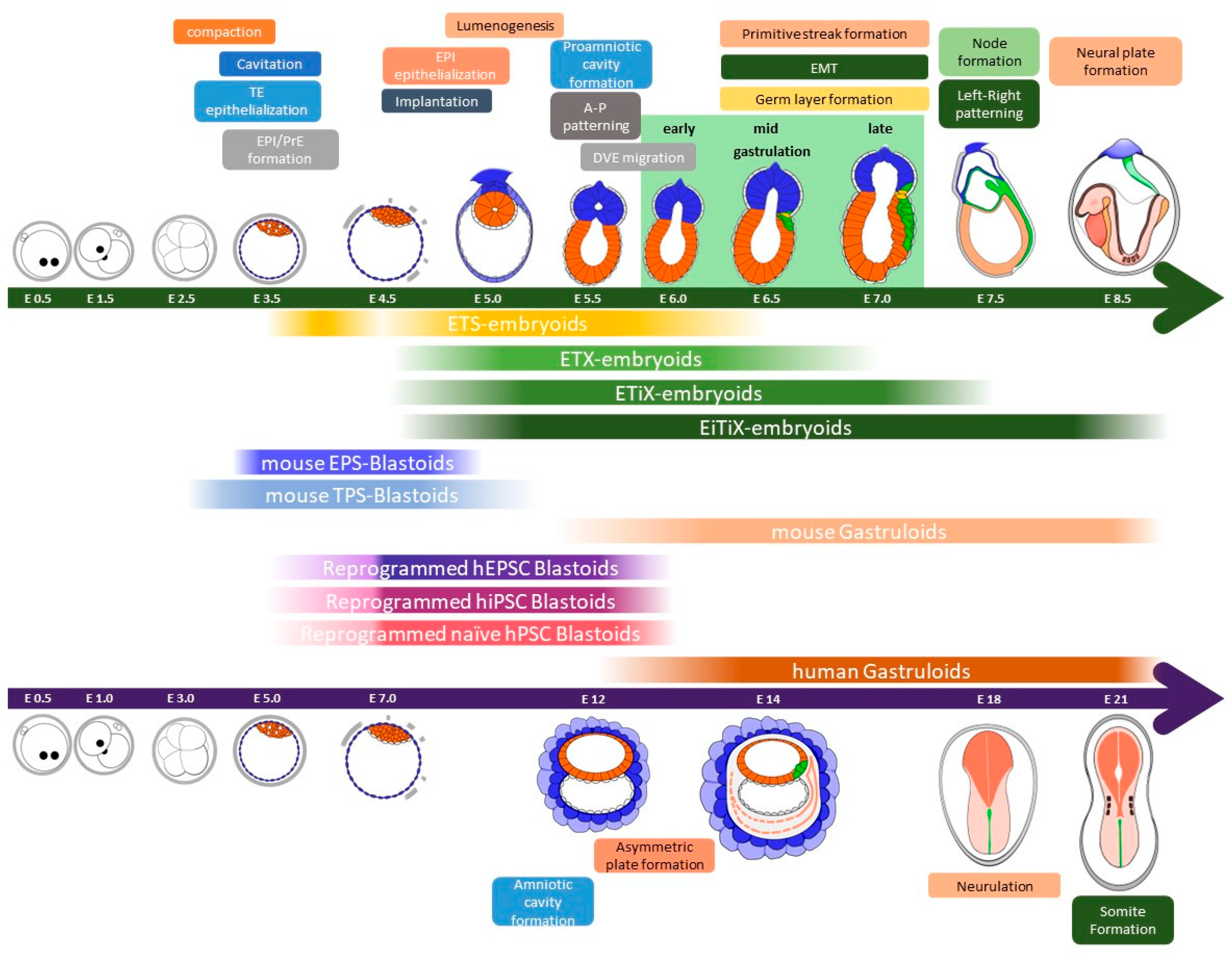
2.2. Peri- and Post-Implantation Embryonic Development
2.3. Gastrulation
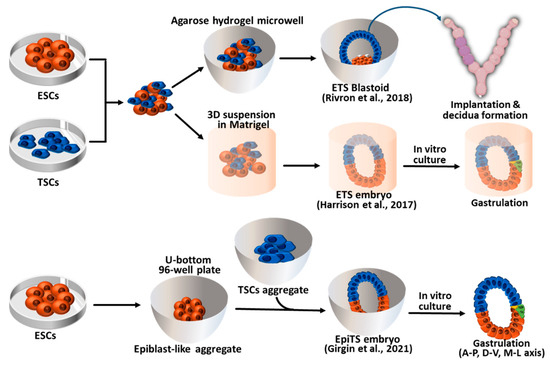
3. Synthetic Embryos Constructed with Mouse ESCs and TSCs (ETS-Embryoids)
Pre-implantation blastocysts consist of three cell layers, namely EPI, TE, and PrE. Each of these cell types can be established as stem cell types in vitro. EPI cells give rise to pluripotent embryonic stem cells (ESCs) in vitro under LIF-Stat3 signaling activation. The other two types of extraembryonic cells, TE and PrE, can also be established as trophoblast stem cells (TSCs) and extraembryonic endoderm cells (XENCs), respectively [52][53][54]. Researchers have attempted to recapitulate pre- and post-implantation embryos using three different types of blastocyst-derived stem cells, specifically ESCs, TSCs, and XENCs. In 2017, an initial trial was conducted by Zernicka-Goetz et al. using only two types of stem cells: ESCs and TSCs. These two stem cell types were combined to generate synthetic embryos which they referred to as “ETS embryos” in Figure 2 [7]. The researchers placed single ESCs and small clumps of TSCs in Matrigel (as a substitute for the PrE) and cultured them in a medium that allowed for the development of ESCs and TSCs. After 96 h of culture, the ETS embryos reached a size of 100 μm × 200 μm, and the number of cells and their morphology were similar to those of natural E5.5 embryos. In the development of ETS embryos, the formation of the proamniotic cavity faithfully recapitulated that of natural embryos. The ESC and TSC cavities formed separately, and at 96 h of ETS embryo development, these cavities merged into a single cavity.
4. Synthetic Embryos Constructed with ESCs, TSCs, and XENCs
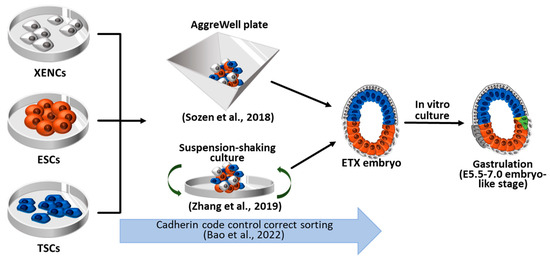
4.1. ETX Embryos Using Wild-Type ESCs, TSCs, and XENCs
4.2. ETiX Embryos Using ESCs Facilitating PrE-Lineage Differentiation
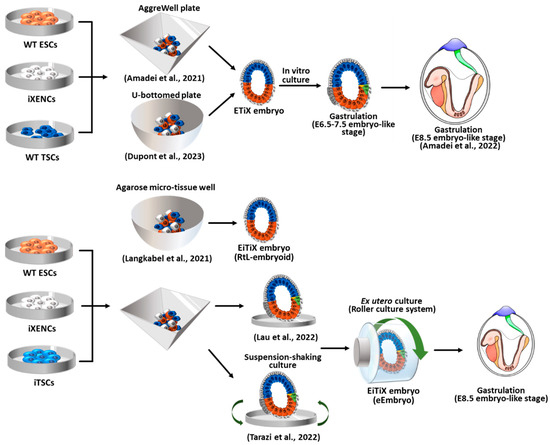
4.3. EiTiX Embryos Constructed with ESCs and Induced TSCs (iTSCs) and Induced XENCs (iXENCs)
5. Blastoid Formation Using Totipotent-Like Stem Cells
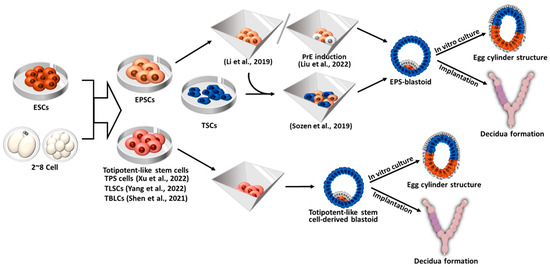
5.1. Blastoid Formation from Expanded Potential Stem Cells (EPSCs) and EPS- and EPST-Blastoids
5.2. Blastoid Formation from Totipotent-Like Stem Cells Other than EPSCs
6. Human Blastoid Formation and In Vitro Implantation Development
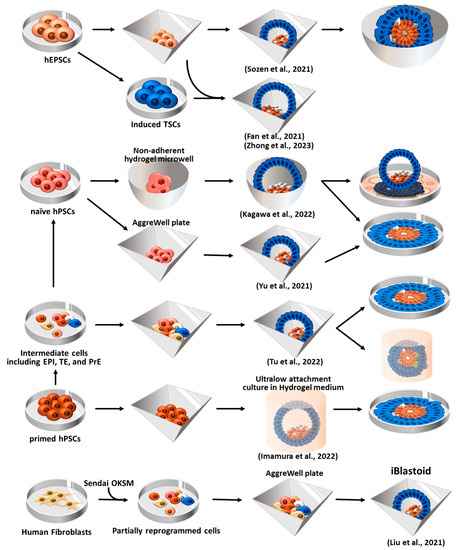
6.1. Blastoid Formation Using Human EPSCs (hEPSCs)
6.2. Human Blastoid Induction via the Reprogramming of Fibroblasts
6.3. Human Blastoid Formation from Primed and Naïve hPSCs
7. Gastruloids
8. Conclusions
This entry is adapted from the peer-reviewed paper 10.3390/ijms241713655
References
- Kojima, Y.; Tam, O.H.; Tam, P.P. Timing of developmental events in the early mouse embryo. Semin. Cell Dev. Biol. 2014, 34, 65–75.
- Ten Berge, D.; Koole, W.; Fuerer, C.; Fish, M.; Eroglu, E.; Nusse, R. Wnt signaling mediates self-organization and axis formation in embryoid bodies. Cell Stem Cell 2008, 3, 508–518.
- Fuchs, C.; Scheinast, M.; Pasteiner, W.; Lagger, S.; Hofner, M.; Hoellrigl, A.; Schultheis, M.; Weitzer, G. Self-organization phenomena in embryonic stem cell-derived embryoid bodies: Axis formation and breaking of symmetry during cardiomyogenesis. Cells Tissues Organs 2012, 195, 377–391.
- Van den Brink, S.C.; Baillie-Johnson, P.; Balayo, T.; Hadjantonakis, A.-K.; Nowotschin, S.; Turner, D.A.; Martinez Arias, A. Symmetry breaking, germ layer specification and axial organisation in aggregates of mouse embryonic stem cells. Development 2014, 141, 4231–4242.
- Warmflash, A.; Sorre, B.; Etoc, F.; Siggia, E.D.; Brivanlou, A.H. A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat. Methods 2014, 11, 847–854.
- Zhang, S.; Chen, T.; Chen, N.; Gao, D.; Shi, B.; Kong, S.; West, R.C.; Yuan, Y.; Zhi, M.; Wei, Q.; et al. Implantation initiation of self-assembled embryo-like structures generated using three types of mouse blastocyst-derived stem cells. Nat. Commun. 2019, 10, 496.
- Harrison, S.E.; Sozen, B.; Christodoulou, N.; Kyprianou, C.; Zernicka-Goetz, M. Assembly of embryonic and extraembryonic stem cells to mimic embryogenesis in vitro. Science 2017, 356, eaal1810.
- Sozen, B.; Amadei, G.; Cox, A.; Wang, R.; Na, E.; Czukiewska, S.; Chappell, L.; Voet, T.; Michel, G.; Jing, N.; et al. Self-assembly of embryonic and two extra-embryonic stem cell types into gastrulating embryo-like structures. Nat. Cell Biol. 2018, 20, 979–989.
- Langkabel, J.; Horne, A.; Bonaguro, L.; Holsten, L.; Hesse, T.; Knaus, A.; Riedel, Y.; Becker, M.; Handler, K.; Elmzzahi, T.; et al. Induction of Rosette-to-Lumen stage embryoids using reprogramming paradigms in ESCs. Nat. Commun. 2021, 12, 7322.
- Girgin, M.U.; Broguiere, N.; Hoehnel, S.; Brandenberg, N.; Mercier, B.; Arias, A.M.; Lutolf, M.P. Bioengineered embryoids mimic post-implantation development in vitro. Nat. Commun. 2021, 12, 5140.
- Amadei, G.; Lau, K.Y.; De Jonghe, J.; Gantner, C.W.; Sozen, B.; Chan, C.; Zhu, M.; Kyprianou, C.; Hollfelder, F.; Zernicka-Goetz, M. Inducible stem-cell-derived embryos capture mouse morphogenetic events in vitro. Dev. Cell 2021, 56, 366–382.e9.
- Amadei, G.; Handford, C.E.; Qiu, C.; De Jonghe, J.; Greenfeld, H.; Tran, M.; Martin, B.K.; Chen, D.Y.; Aguilera-Castrejon, A.; Hanna, J.H.; et al. Embryo model completes gastrulation to neurulation and organogenesis. Nature 2022, 610, 143–153.
- Dupont, C.; Schäffers, O.J.; Tan, B.F.; Merzouk, S.; Bindels, E.M.; Zwijsen, A.; Huylebroeck, D.; Gribnau, J. Efficient generation of ETX embryoids that recapitulate the entire window of murine egg cylinder development. Sci. Adv. 2023, 9, eadd2913.
- Aguilera-Castrejon, A.; Oldak, B.; Shani, T.; Ghanem, N.; Itzkovich, C.; Slomovich, S.; Tarazi, S.; Bayerl, J.; Chugaeva, V.; Ayyash, M.; et al. Ex utero mouse embryogenesis from pre-gastrulation to late organogenesis. Nature 2021, 593, 119–124.
- Lau, K.Y.C.; Rubinstein, H.; Gantner, C.W.; Hadas, R.; Amadei, G.; Stelzer, Y.; Zernicka-Goetz, M. Mouse embryo model derived exclusively from embryonic stem cells undergoes neurulation and heart development. Cell Stem Cell 2022, 29, 1445–1458.e8.
- Tarazi, S.; Aguilera-Castrejon, A.; Joubran, C.; Ghanem, N.; Ashouokhi, S.; Roncato, F.; Wildschutz, E.; Haddad, M.; Oldak, B.; Gomez-Cesar, E.; et al. Post-gastrulation synthetic embryos generated ex utero from mouse naive ESCs. Cell 2022, 185, 3290–3306.e25.
- Bedzhov, I.; Leung, C.Y.; Bialecka, M.; Zernicka-Goetz, M. In vitro culture of mouse blastocysts beyond the implantation stages. Nat. Protoc. 2014, 9, 2732–2739.
- Xu, Y.; Zhao, J.; Ren, Y.; Wang, X.; Lyu, Y.; Xie, B.; Sun, Y.; Yuan, X.; Liu, H.; Yang, W.; et al. Derivation of totipotent-like stem cells with blastocyst-like structure forming potential. Cell Res. 2022, 32, 513–529.
- Yang, M.; Yu, H.; Yu, X.; Liang, S.; Hu, Y.; Luo, Y.; Izsvak, Z.; Sun, C.; Wang, J. Chemical-induced chromatin remodeling reprograms mouse ESCs to totipotent-like stem cells. Cell Stem Cell 2022, 29, 400–418.e13.
- Shen, H.; Yang, M.; Li, S.; Zhang, J.; Peng, B.; Wang, C.; Chang, Z.; Ong, J.; Du, P. Mouse totipotent stem cells captured and maintained through spliceosomal repression. Cell 2021, 184, 2843–2859.e20.
- Zhang, P.; Zhai, X.; Huang, B.; Sun, S.; Wang, W.; Zhang, M. Highly efficient generation of blastocyst-like structures from spliceosomes-repressed mouse totipotent blastomere-like cells. Sci. China Life Sci. 2023, 66, 423–435.
- Cossec, J.-C.; Traboulsi, T.; Sart, S.; Loe-Mie, Y.; Guthmann, M.; Hendriks, I.A.; Theurillat, I.; Nielsen, M.L.; Torres-Padilla, M.-E.; Baroud, C.N. Transient suppression of SUMOylation in embryonic stem cells generates embryo-like structures. Cell Rep. 2023, 42, 112380.
- Jenkinson, E.; Wilson, I. In vitro support system for the study of blastocyst differentiation in the mouse. Nature 1970, 228, 776–778.
- Condic, M.L. Totipotency: What it is and what it is not. Stem Cells Dev. 2014, 23, 796–812.
- Maemura, M.; Taketsuru, H.; Nakajima, Y.; Shao, R.; Kakihara, A.; Nogami, J.; Ohkawa, Y.; Tsukada, Y.I. Totipotency of mouse zygotes extends to single blastomeres of embryos at the four-cell stage. Sci. Rep. 2021, 11, 11167.
- Johnson, W.H.; Loskutoff, N.M.; Plante, Y.; Betteridge, K.J. Production of four identical calves by the separation of blastomeres from an in vitro derived four-cell embryo. Vet. Rec. 1995, 137, 15–16.
- Calarco, P.G.; Brown, E.H. An ultrastructural and cytological study of preimplantation development of the mouse. J. Exp. Zool. 1969, 171, 253–283.
- Ducibella, T.; Ukena, T.; Karnovsky, M.; Anderson, E. Changes in cell surface and cortical cytoplasmic organization during early embryogenesis in the preimplantation mouse embryo. J. Cell Biol. 1977, 74, 153–167.
- White, M.D.; Bissiere, S.; Alvarez, Y.D.; Plachta, N. Mouse Embryo Compaction. Curr. Top. Dev. Biol. 2016, 120, 235–258.
- Arnold, S.J.; Robertson, E.J. Making a commitment: Cell lineage allocation and axis patterning in the early mouse embryo. Nat. Rev. Mol. Cell Biol. 2009, 10, 91–103.
- Chazaud, C.; Yamanaka, Y.; Pawson, T.; Rossant, J. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev. Cell 2006, 10, 615–624.
- Niwa, H.; Toyooka, Y.; Shimosato, D.; Strumpf, D.; Takahashi, K.; Yagi, R.; Rossant, J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 2005, 123, 917–929.
- Varelas, X. The Hippo pathway effectors TAZ and YAP in development, homeostasis and disease. Development 2014, 141, 1614–1626.
- Nishioka, N.; Inoue, K.; Adachi, K.; Kiyonari, H.; Ota, M.; Ralston, A.; Yabuta, N.; Hirahara, S.; Stephenson, R.O.; Ogonuki, N.; et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell 2009, 16, 398–410.
- Bedzhov, I.; Zernicka-Goetz, M. Self-organizing properties of mouse pluripotent cells initiate morphogenesis upon implantation. Cell 2014, 156, 1032–1044.
- Varlet, I.; Collignon, J.; Robertson, E.J. nodal expression in the primitive endoderm is required for specification of the anterior axis during mouse gastrulation. Development 1997, 124, 1033–1044.
- Papanayotou, C.; Benhaddou, A.; Camus, A.; Perea-Gomez, A.; Jouneau, A.; Mezger, V.; Langa, F.; Ott, S.; Sabéran-Djoneidi, D.; Collignon, J. A novel nodal enhancer dependent on pluripotency factors and smad2/3 signaling conditions a regulatory switch during epiblast maturation. PLoS Biol. 2014, 12, e1001890.
- Senft, A.D.; Bikoff, E.K.; Robertson, E.J.; Costello, I. Genetic dissection of Nodal and Bmp signalling requirements during primordial germ cell development in mouse. Nat. Commun. 2019, 10, 1089.
- Brennan, J.; Lu, C.C.; Norris, D.P.; Rodriguez, T.A.; Beddington, R.S.P.; Robertson, E.J. Nodal signalling in the epiblast patterns the early mouse embryo. Nature 2001, 411, 965–969.
- Ciruna, B.; Rossant, J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev. Cell 2001, 1, 37–49.
- Ciruna, B.G.; Schwartz, L.; Harpal, K.; Yamaguchi, T.P.; Rossant, J. Chimeric analysis of fibroblast growth factor receptor-1 (Fgfr1) function: A role for FGFR1 in morphogenetic movement through the primitive streak. Development 1997, 124, 2829–2841.
- Huelsken, J.; Vogel, R.; Brinkmann, V.; Erdmann, B.; Birchmeier, C.; Birchmeier, W. Requirement for beta-catenin in anterior-posterior axis formation in mice. J. Cell Biol. 2000, 148, 567–578.
- Mishina, Y.; Suzuki, A.; Ueno, N.; Behringer, R.R. Bmpr encodes a type I bone morphogenetic protein receptor that is essential for gastrulation during mouse embryogenesis. Genes Dev. 1995, 9, 3027–3037.
- Liu, P.; Wakamiya, M.; Shea, M.J.; Albrecht, U.; Behringer, R.R.; Bradley, A. Requirement for Wnt3 in vertebrate axis formation. Nat. Genet. 1999, 22, 361–365.
- Kinder, S.J.; Tsang, T.E.; Wakamiya, M.; Sasaki, H.; Behringer, R.R.; Nagy, A.; Tam, P.P. The organizer of the mouse gastrula is composed of a dynamic population of progenitor cells for the axial mesoderm. Development 2001, 128, 3623–3634.
- Sulik, K.; Dehart, D.B.; Iangaki, T.; Carson, J.L.; Vrablic, T.; Gesteland, K.; Schoenwolf, G.C. Morphogenesis of the murine node and notochordal plate. Dev. Dyn. 1994, 201, 260–278.
- Yamanaka, Y.; Tamplin, O.J.; Beckers, A.; Gossler, A.; Rossant, J. Live imaging and genetic analysis of mouse notochord formation reveals regional morphogenetic mechanisms. Dev. Cell 2007, 13, 884–896.
- Pituello, F. Neuronal specification: Generating diversity in the spinal cord. Curr. Biol. 1997, 7, R701–R704.
- Lawson, K.A. Fate mapping the mouse embryo. Int. J. Dev. Biol. 1999, 43, 773–775.
- Di-Gregorio, A.; Sancho, M.; Stuckey, D.W.; Crompton, L.A.; Godwin, J.; Mishina, Y.; Rodriguez, T.A. BMP signalling inhibits premature neural differentiation in the mouse embryo. Development 2007, 134, 3359–3369.
- Camus, A.; Perea-Gomez, A.; Moreau, A.; Collignon, J. Absence of Nodal signaling promotes precocious neural differentiation in the mouse embryo. Dev. Biol. 2006, 295, 743–755.
- Tanaka, S.; Kunath, T.; Hadjantonakis, A.K.; Nagy, A.; Rossant, J. Promotion of trophoblast stem cell proliferation by FGF4. Science 1998, 282, 2072–2075.
- Nichols, J.; Zevnik, B.; Anastassiadis, K.; Niwa, H.; Klewe-Nebenius, D.; Chambers, I.; Schöler, H.; Smith, A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 1998, 95, 379–391.
- Ying, Q.-L.; Wray, J.; Nichols, J.; Batlle-Morera, L.; Doble, B.; Woodgett, J.; Cohen, P.; Smith, A. The ground state of embryonic stem cell self-renewal. Nature 2008, 453, 519–523.
- Rivron, N.C.; Frias-Aldeguer, J.; Vrij, E.J.; Boisset, J.C.; Korving, J.; Vivie, J.; Truckenmuller, R.K.; van Oudenaarden, A.; van Blitterswijk, C.A.; Geijsen, N. Blastocyst-like structures generated solely from stem cells. Nature 2018, 557, 106–111.
- Kunath, T.; Arnaud, D.; Uy, G.D.; Okamoto, I.; Chureau, C.; Yamanaka, Y.; Heard, E.; Gardner, R.L.; Avner, P.; Rossant, J. Imprinted X-inactivation in extra-embryonic endoderm cell lines from mouse blastocysts. Development 2005, 132, 1649–1661.
- Enders, A.C.; Given, R.L.; Schlafke, S. Differentiation and migration of endoderm in the rat and mouse at implantation. Anat. Rec. 1978, 190, 65–77.
- Bao, M.; Cornwall-Scoones, J.; Sanchez-Vasquez, E.; Cox, A.L.; Chen, D.-Y.; De Jonghe, J.; Shadkhoo, S.; Hollfelder, F.; Thomson, M.; Glover, D.M.; et al. Stem cell-derived synthetic embryos self-assemble by exploiting cadherin codes and cortical tension. Nat. Cell Biol. 2022, 24, 1341–1349.
- Tam, P.P.; Beddington, R.S. Establishment and organization of germ layers in the gastrulating mouse embryo. Ciba Found. Symp. 1992, 165, 27–41; discussion 42–49.
- Viotti, M.; Nowotschin, S.; Hadjantonakis, A.K. Afp::mCherry, a red fluorescent transgenic reporter of the mouse visceral endoderm. Genesis 2011, 49, 124–133.
- Perea-Gomez, A.; Meilhac, S.M.; Piotrowska-Nitsche, K.; Gray, D.; Collignon, J.; Zernicka-Goetz, M. Regionalisation of the mouse visceral endoderm as the blastocyst transforms into the egg cylinder. BMC Dev. Biol. 2007, 7, 96.
- Li, S.; Edgar, D.; Fässler, R.; Wadsworth, W.; Yurchenco, P.D. The role of laminin in embryonic cell polarization and tissue organization. Dev. Cell 2003, 4, 613–624.
- Paca, A.; Séguin, C.A.; Clements, M.; Ryczko, M.; Rossant, J.; Rodriguez, T.A.; Kunath, T. BMP signaling induces visceral endoderm differentiation of XEN cells and parietal endoderm. Dev. Biol. 2012, 361, 90–102.
- Moerkamp, A.T.; Paca, A.; Goumans, M.J.; Kunath, T.; Kruithof, B.P.; Kruithof-de Julio, M. Extraembryonic endoderm cells as a model of endoderm development. Dev. Growth Differ. 2013, 55, 301–308.
- Shimosato, D.; Shiki, M.; Niwa, H. Extra-embryonic endoderm cells derived from ES cells induced by GATA factors acquire the character of XEN cells. BMC Dev. Biol. 2007, 7, 80.
- Schröter, C.; Rué, P.; Mackenzie, J.P.; Martinez Arias, A. FGF/MAPK signaling sets the switching threshold of a bistable circuit controlling cell fate decisions in embryonic stem cells. Development 2015, 142, 4205–4216.
- Tosic, J.; Kim, G.-J.; Pavlovic, M.; Schröder, C.M.; Mersiowsky, S.-L.; Barg, M.; Hofherr, A.; Probst, S.; Köttgen, M.; Hein, L. Eomes and Brachyury control pluripotency exit and germ-layer segregation by changing the chromatin state. Nature 2019, 21, 1518–1531.
- Ramkumar, N.; Omelchenko, T.; Silva-Gagliardi, N.F.; McGlade, C.J.; Wijnholds, J.; Anderson, K.V. Crumbs2 promotes cell ingression during the epithelial-to-mesenchymal transition at gastrulation. Nat. Cell Biol. 2016, 18, 1281–1291.
- Nowotschin, S.; Setty, M.; Kuo, Y.-Y.; Liu, V.; Garg, V.; Sharma, R.; Simon, C.S.; Saiz, N.; Gardner, R.; Boutet, S.C. The emergent landscape of the mouse gut endoderm at single-cell resolution. Nature 2019, 569, 361–367.
- Tanaka, Y.; Hayashi, M.; Kubota, Y.; Nagai, H.; Sheng, G.; Nishikawa, S.-I.; Samokhvalov, I.M. Early ontogenic origin of the hematopoietic stem cell lineage. Proc. Natl. Acad. Sci. USA 2012, 109, 4515–4520.
- Tanaka, Y.; Sanchez, V.; Takata, N.; Yokomizo, T.; Yamanaka, Y.; Kataoka, H.; Hoppe, P.S.; Schroeder, T.; Nishikawa, S.-I. Circulation-independent differentiation pathway from extraembryonic mesoderm toward hematopoietic stem cells via hemogenic angioblasts. Cell Rep. 2014, 8, 31–39.
- Ross, C.; Boroviak, T.E. Origin and function of the yolk sac in primate embryogenesis. Nat. Commun. 2020, 11, 3760.
- Frankenberg, S.; Gerbe, F.; Bessonnard, S.; Belville, C.; Pouchin, P.; Bardot, O.; Chazaud, C. Primitive endoderm differentiates via a three-step mechanism involving Nanog and RTK signaling. Dev. Cell 2011, 21, 1005–1013.
- Saitou, M.; Yamaji, M. Germ cell specification in mice: Signaling, transcription regulation, and epigenetic consequences. Reproduction 2010, 139, 931.
- Pereira, P.N.; Dobreva, M.P.; Graham, L.; Huylebroeck, D.; Lawson, K.A.; Zwijsen, A. Amnion formation in the mouse embryo: The single amniochorionic fold model. BMC Dev. Biol. 2011, 11, 48.
- Huber, T.L.; Kouskoff, V.; Fehling, H.J.; Palis, J.; Keller, G. Haemangioblast commitment is initiated in the primitive streak of the mouse embryo. Nature 2004, 432, 625–630.
- Bergiers, I.; Andrews, T.; Vargel Bölükbaşı, Ö.; Buness, A.; Janosz, E.; Lopez-Anguita, N.; Ganter, K.; Kosim, K.; Celen, C.; Itır Perçin, G. Single-cell transcriptomics reveals a new dynamical function of transcription factors during embryonic hematopoiesis. Elife 2018, 7, e29312.
- Home, P.; Ray, S.; Dutta, D.; Bronshteyn, I.; Larson, M.; Paul, S. GATA3 is selectively expressed in the trophectoderm of peri-implantation embryo and directly regulates Cdx2 gene expression. J. Biol. Chem. 2009, 284, 28729–28737.
- Kresoja-Rakic, J.; Santoro, R. Nucleolus and rRNA gene chromatin in early embryo development. Trends Genet. 2019, 35, 868–879.
- Li, R.; Zhong, C.; Yu, Y.; Liu, H.; Sakurai, M.; Yu, L.; Min, Z.; Shi, L.; Wei, Y.; Takahashi, Y.; et al. Generation of Blastocyst-like Structures from Mouse Embryonic and Adult Cell Cultures. Cell 2019, 179, 687–702.e18.
- Liu, K.; Xu, X.; Bai, D.; Li, Y.; Zhang, Y.; Jia, Y.; Guo, M.; Han, X.; Liu, Y.; Sheng, Y.; et al. Bilineage embryo-like structure from EPS cells can produce live mice with tetraploid trophectoderm. Protein Cell 2022, 14, 262–278.
- Sozen, B.; Cox, A.L.; De Jonghe, J.; Bao, M.; Hollfelder, F.; Glover, D.M.; Zernicka-Goetz, M. Self-Organization of Mouse Stem Cells into an Extended Potential Blastoid. Dev. Cell 2019, 51, 698–712.e8.
- Yang, J.; Ryan, D.J.; Wang, W.; Tsang, J.C.-H.; Lan, G.; Masaki, H.; Gao, X.; Antunes, L.; Yu, Y.; Zhu, Z.; et al. Establishment of mouse expanded potential stem cells. Nature 2017, 550, 393–397.
- Posfai, E.; Schell, J.P.; Janiszewski, A.; Rovic, I.; Murray, A.; Bradshaw, B.; Yamakawa, T.; Pardon, T.; El Bakkali, M.; Talon, I. Evaluating totipotency using criteria of increasing stringency. Nat. Cell Biol. 2021, 23, 49–60.
- Suwińska, A. Preimplantation mouse embryo: Developmental fate and potency of blastomeres. Mouse Dev. Oocyte Stem. Cells 2012, 55, 141–163.
- Genet, M.; Torres-Padilla, M.-E. The molecular and cellular features of 2-cell-like cells: A reference guide. Development 2020, 147, dev189688.
- Fu, J.; Warmflash, A.; Lutolf, M.P. Stem-cell-based embryo models for fundamental research and translation. Nat. Mater. 2021, 20, 132–144.
- Shahbazi, M.N.; Jedrusik, A.; Vuoristo, S.; Recher, G.; Hupalowska, A.; Bolton, V.; Fogarty, N.N.M.; Campbell, A.; Devito, L.; Ilic, D.; et al. Self-organization of the human embryo in the absence of maternal tissues. Nat. Cell Biol. 2016, 18, 700–708.
- Xiang, L.; Yin, Y.; Zheng, Y.; Ma, Y.; Li, Y.; Zhao, Z.; Guo, J.; Ai, Z.; Niu, Y.; Duan, K. A developmental landscape of 3D-cultured human pre-gastrulation embryos. Nature 2020, 577, 537–542.
- Deglincerti, A.; Croft, G.F.; Pietila, L.N.; Zernicka-Goetz, M.; Siggia, E.D.; Brivanlou, A.H. Self-organization of the in vitro attached human embryo. Nature 2016, 533, 251–254.
- Liu, X.; Tan, J.P.; Schroder, J.; Aberkane, A.; Ouyang, J.F.; Mohenska, M.; Lim, S.M.; Sun, Y.B.Y.; Chen, J.; Sun, G.; et al. Modelling human blastocysts by reprogramming fibroblasts into iBlastoids. Nature 2021, 591, 627–632.
- Sozen, B.; Jorgensen, V.; Weatherbee, B.A.T.; Chen, S.; Zhu, M.; Zernicka-Goetz, M. Reconstructing aspects of human embryogenesis with pluripotent stem cells. Nat. Commun. 2021, 12, 5550.
- Zhong, K.; Luo, Y.-X.; Li, D.; Min, Z.-Y.; Fan, Y.; Yu, Y. Generation of blastoids from human parthenogenetic stem cells. Life Med. 2023, 2, lnad006.
- Yu, L.; Wei, Y.; Duan, J.; Schmitz, D.A.; Sakurai, M.; Wang, L.; Wang, K.; Zhao, S.; Hon, G.C.; Wu, J. Blastocyst-like structures generated from human pluripotent stem cells. Nature 2021, 591, 620–626.
- Fan, Y.; Min, Z.; Alsolami, S.; Ma, Z.; Zhang, E.; Chen, W.; Zhong, K.; Pei, W.; Kang, X.; Zhang, P. Generation of human blastocyst-like structures from pluripotent stem cells. Cell Discov. 2021, 7, 81.
- Kagawa, H.; Javali, A.; Khoei, H.H.; Sommer, T.M.; Sestini, G.; Novatchkova, M.; Scholte Op Reimer, Y.; Castel, G.; Bruneau, A.; Maenhoudt, N.; et al. Human blastoids model blastocyst development and implantation. Nature 2022, 601, 600–605.
- Tu, Z.; Bi, Y.; Zhu, X.; Liu, W.; Hu, J.; Wu, L.; Mao, T.; Zhou, J.; Wang, H.; Wang, H. Modeling human pregastrulation development by 3D culture of blastoids generated from primed-to-naïve transitioning intermediates. Protein Cell 2023, 14, 337–349.
- Imamura, S.; Wen, X.; Terada, S.; Yamamoto, A.; Mutsuda-Zapater, K.; Sawada, K.; Yoshimoto, K.; Tanaka, M.; Kamei, K.-i. Human blastoid from primed human embryonic stem cells. bioRxiv 2022.
- Okae, H.; Toh, H.; Sato, T.; Hiura, H.; Takahashi, S.; Shirane, K.; Kabayama, Y.; Suyama, M.; Sasaki, H.; Arima, T. Derivation of human trophoblast stem cells. Cell Stem Cell 2018, 22, 50–63.e6.
- Liu, X.; Ouyang, J.F.; Rossello, F.J.; Tan, J.P.; Davidson, K.C.; Valdes, D.S.; Schröder, J.; Sun, Y.B.; Chen, J.; Knaupp, A.S. Reprogramming roadmap reveals route to human induced trophoblast stem cells. Nature 2020, 586, 101–107.
- Dong, C.; Beltcheva, M.; Gontarz, P.; Zhang, B.; Popli, P.; Fischer, L.A.; Khan, S.A.; Park, K.M.; Yoon, E.J.; Xing, X.; et al. Derivation of trophoblast stem cells from naïve human pluripotent stem cells. elife 2020, 9, e52504.
- Dodsworth, B.T.; Hatje, K.; Rostovskaya, M.; Flynn, R.; Meyer, C.A.; Cowley, S.A. Profiling of naïve and primed human pluripotent stem cells reveals state-associated miRNAs. Sci. Rep. 2020, 10, 10542.
- Guo, G.; Stirparo, G.G.; Strawbridge, S.E.; Spindlow, D.; Yang, J.; Clarke, J.; Dattani, A.; Yanagida, A.; Li, M.A.; Myers, S.; et al. Human naive epiblast cells possess unrestricted lineage potential. Cell Stem Cell 2021, 28, 1040–1056.e6.
- Linneberg-Agerholm, M.; Wong, Y.F.; Romero Herrera, J.A.; Monteiro, R.S.; Anderson, K.G.; Brickman, J.M. Naïve human pluripotent stem cells respond to Wnt, Nodal and LIF signalling to produce expandable naïve extra-embryonic endoderm. Development 2019, 146, dev180620.
- Theunissen, T.W.; Powell, B.E.; Wang, H.; Mitalipova, M.; Faddah, D.A.; Reddy, J.; Fan, Z.P.; Maetzel, D.; Ganz, K.; Shi, L. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell 2014, 15, 471–487.
- van den Brink, S.C.; Alemany, A.; van Batenburg, V.; Moris, N.; Blotenburg, M.; Vivié, J.; Baillie-Johnson, P.; Nichols, J.; Sonnen, K.F.; Martinez Arias, A.; et al. Single-cell and spatial transcriptomics reveal somitogenesis in gastruloids. Nature 2020, 582, 405–409.
- Veenvliet, J.V.; Bolondi, A.; Kretzmer, H.; Haut, L.; Scholze-Wittler, M.; Schifferl, D.; Koch, F.; Guignard, L.; Kumar, A.S.; Pustet, M. Mouse embryonic stem cells self-organize into trunk-like structures with neural tube and somites. Science 2020, 370, eaba4937.
