Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Liver fibrosis represents the reversible pathological process with the feature of the over-accumulation of extracellular matrix (ECM) proteins within the liver, which results in the deposition of fibrotic tissues and liver dysfunction. Circular noncoding RNAs (CircRNAs) have the characteristic closed loop structures, which show high resistance to exonuclease RNase, making them far more stable and recalcitrant against degradation. CircRNAs increase target gene levels by playing the role of a microRNA (miRNA) sponge.
- circRNA
- liver fibrosis
- ceRNA
1. Introduction
Various chronic irritations can damage the liver, which thus causes inflammation and necrosis of hepatocytes, activates the hepatic stellate cells, and produces the significant accumulation of extracellular matrix (ECM) that is extremely rich in type I collagen and other collagens [1], eventually resulting in liver fibrosis. Liver fibrosis, a common pathological change in various chronic liver diseases [2], presents as a precursor to liver cirrhosis, and it finally develops into liver failure and hepatocellular carcinoma (HCC) [3]. Thus, it is critical to slow down, or ideally reverse liver fibrosis progression. According to recent articles, many types of CircRNAs are related to liver fibrosis occurrence and development. For instance, CircPWWP2A has been suggested to enhance hepatic stellate cells (HSCs) growth and activation and liver fibrosis by acting as the sponge of miR-203 and miR-223, which then up-regulate Fstl1 and Tlr4, respectively [4]. However, biological functions and molecular mechanisms underlying CircRNAs in hepatic fibrosis (HF), particularly in relation to the activation of HSCs, remain ambiguous and require further investigations.
2. Mechanism of CircRNAs in Liver Fibrosis
Many CircRNAs have been widely suggested to be related to promoting or inhibiting the development and progression of liver fibrosis; therefore, they may serve as valuable biomarkers for monitoring liver fibrosis development (Figure 1) [4][5][6][7]. For example, based on experimental data from Zhou et al., CircRNAs expression profiles within liver fibrosis tissues were remarkably altered. Some of the CircRNAs with differential expression are closely related to physiological processes, such as stellate cell activation, macrophage inflammation, and oxidative stress damage. The target genes of CircRNAs are primarily involved in pathways, such as HIPPO, TGF-β1/Smads, vascular endothelial growth factor (VEGF), and RAP1 [8]. Meanwhile, Chen et al. used the CircRNA microarray for investigating CircRNA expression profiles of irradiated HSCs from HCC. According to their results, there were 179 CircRNAs showing up-regulation, whereas 630 displayed down-regulation. Bioinformatics analysis implied that the abnormal expression of CircRNAs was possibly related to cell responses to radiation and the biological process of liver fibrosis [9]. Therefore, elucidating the relationships between the aberrant expression of CircRNAs within HSCs with liver fibrosis is of crucial importance.
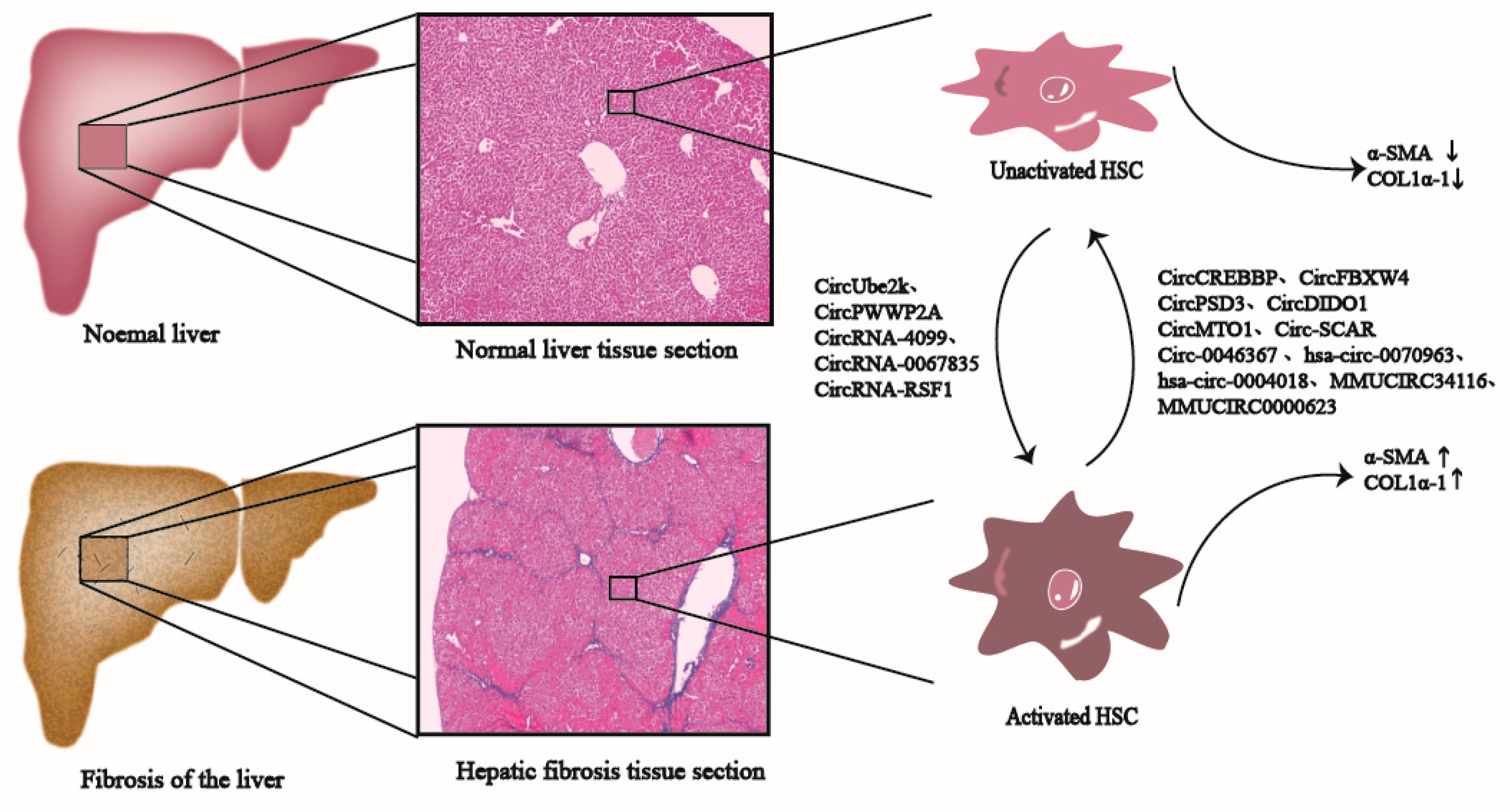
Figure 1. CircRNAs participate in the liver fibrosis process. CircRNAs, such as CircUbe2k, CircPWWP2A, and CircRNA-4099 promote the activation of HSCs in normal liver tissues, generate pro-fibrosis factors such as α-SMA and Col1α1, and ultimately promote the development of liver fibrosis; CircRNAs, such as CircCREBBP, CircFBXW4, and CircPSD3 inhibit the activation of HSCs, reduce the production of pro-fibrosis factors such as α-SMA and Col1α1, and eventually inhibit liver fibrosis.
2.1. CircRNAs Are Involved in Inhibiting Liver Fibrosis
2.1.1. CircRNAs Are Related to Liver Fibrosis through Serving as RNA Scaffolds
CircRNA SCAR
After analyzing CircRNA expression patterns in primary liver fibroblasts collected in NASH cirrhosis cases, Zhao et al. uncovered that 15 and 11 CircRNAs displayed up-regulation and down-regulation, separately, within NASH fibroblasts, including four mitochondria-specific CircRNAs. Next, those four mitochondria-specific CircRNAs were introduced into the NASH fibroblasts by synthesizing the mitochondrial-targeted nanoparticles (mito-NP). It was found that only the hsa_circ_0089762 overexpression (steatohepatitis-associated CircRNA ATP5B regulator [SCAR]) significantly reduced the level of cytosolic ROS (cROS). This clearly inhibited the contractility, collagen generation, α-SMA expression, and cytokine secretion in NASH fibroblasts, thus alleviating the fibrotic phenotype [10].
The main mechanism involves the direct binding of CircRNA SCAR to ATP5B within the mPTP complex, which closes mPTP by blocking CypD-MPTP interactions and reducing the generation and release of mROS, thereby inhibiting the activation of fibroblasts. In the NASH liver fibroblasts, lipid overload can up-regulate the endoplasmic reticulum stress protein CHOP. This inhibits PGC-1a (peroxisome proliferator-activated receptor gamma, coactivator 1 alpha), then suppresses the transcription of CircRNA SCAR, and eventually promotes the activation of fibroblasts [10]. Researchers have given mito-NP that contains CircRNA SCAR overexpression vectors (or empty vectors) to mice that were fed with a high-fat diet through intravenous administration. According to their observations, the increased CircRNA SCAR significantly inhibits the mPTP opening, cROS release, and collagen contraction of mouse fibroblasts. This obviously improves glucose and insulin tolerance and halts weight gain, while reducing fibrosis and macrophage infiltration in the mouse livers. In clinical practice, CircRNA SCAR is related to steatosis-to-NASH progression [10]; in this regard, a mitochondrial CircRNA SCAR modulates lipid metabolism and may be the anti-NASH therapeutic target.
2.1.2. CircRNAs Are Related to Liver Fibrosis by Playing a Role of miRNA-Binding Cernas
CircCREBBP
As a highly conserved transcription coactivator, CREBBP possesses histone acetyltransferase activities [11]. However, its mutation or deficiency is frequently detected in human cancers, which suggests that it may serve as a tumor suppressor in pathophysiological processes [12][13]. CircCREBBP (hsa_circ_0007673, mmu_circ_0006288) originates from the backsplicing of the CREBBP exons [6] located in 16p13.3 [14].
Through conducting high-throughput sequencing, Yang et al. detected CircRNAs expression patterns within the fibrotic liver. As a result, there were 103 CircRNAs showing differential expression, including 18 with up-regulation and 85 with down-regulation within HF tissues [6]. Among them, CircCREBBP exhibited significant downregulation among HF mice. As revealed by the AAV8-induced CircCREBBP overexpression within mice, CircCREBBP significantly suppresses HSCs growth and activation because it blocks the cell cycle, reduces the transdifferentiation of myofibroblasts, decreases collagen deposition, and inhibits fibrogenic factor levels, thereby preventing the worsening of CCl4-induced HF [6]. From the mechanism perspective, CircCREBBP is the sponge for hsa-miR-1291, which up-regulates LEFTY2, thus alleviating liver fibrosis within TGF-β1-treated LX-2 cells, primary HSCs, and liver tissues from CCl4-induced HF mice, and liver fibrosis patients via the hsa-miR-1291/LEFTY2 axis [6].
CircFBXW4
CircFBXW4 (Mm9_circ_000338) is located on chromosome 19: 45705354-45715011 (510 nt) and consists of four exons (exon 2–5) at FBXW4 (the F box and WD 40 domain-containing protein 4) gene locus [5]. Chen et al. analyzed CircRNAs levels within HSCs in chronic liver fibrosis mice by CircRNAs-seq and reported that CircFBXW4 expression was significantly down-regulated during liver fibrogenesis, and it facilitated the HF recovery process [5]. The lower levels of CircFBXW4 are also present within peripheral blood from HF cases, in contrast to normal subjects. The expression of CircFBXW7 is consistently down-regulated within acute liver injury, CCl4, and BDL-induced HF mice. Further, the in vivo overexpression of CircFBXW4 by liver-specific administration of pHBAAV-circFBXW4 decreases collagen deposition and myofibroblast transdifferentiation, mitigates injuries to mouse liver fibrogenesis, and avoids inflammatory response, which reveals that CircFBXW4 resists against fibrosis in HF [5]. Moreover, the in vitro CircFBXW4 overexpression suppresses LX-2 cell activation, inhibits cell growth as well as DNA synthesis, causes cell cycle arrest, and ultimately induces LX-2 cell apoptosis. Eventually, as revealed from the luciferase reporter, fluorescence in situ hybridization and RNA pull-down assays conducted by Chen et al., CircFBXW4 bound to miR-18b-3p to modulate FBXW7 level as the miRNA sponge [5]. In view of its crucial roles in liver fibrosis and HSC activation, CircFBXW4 may be the candidate marker used to diagnose and treat the disease.
CircPSD3
It is found that CircPSD3, which is derived from exons 4–8 of pleckstrin and the Sec7 domain that contains a three (PSD3) gene locus, markedly decreases within primary HSCs together with the liver tissues in CCl4-mediated HF mice. The in vivo CircPSD3 overexpression induced by AAV8-circPSD3 injection inhibits HSCs activation and proliferation and alleviates CCl4-induced HF, which can be evidenced by the decreased serum aspartate aminotransferase (AST) and alanine aminotransferase (ALT) levels, collagen deposition, liver hydroxyproline content, along with pro-inflammatory cytokine and pro-fibrogenic gene levels [15]. The in vitro CircPSD3 overexpression significantly decreased a-SMA and Col1α1 expression at mRNA and protein levels, while inhibiting HSC growth and activation. CircPSD3 is the mechanical sponge for miR-92b-3p, which can later promote Smad7 expression. Specifically, CircPSD3 alleviates the formation of liver fibrosis via miR-92b-3p/Smad7 axis, which suggests that CircPSD3 may be a potential biomarker for HF [15].
Hsa_circ_0070963
Hsa_circ_0070963 shows decreased expression within mice with CCl4-induced liver fibrosis, while the restoration of hsa_circ_0070963 expression can stop HSC activation and decrease type I collagen and α-SMA expression, both in vivo and in vitro [16]. In addition, as revealed by a luciferase reporter assay and rescue experimenthsa_circ_0070963 is confirmed to sponge miR-223-3p to inhibit HSC activation during liver fibrosis through regulating miR-223-3p and LEMD3 [16], while LEMD3 (called MAN1 as well), as the inner nuclear membrane protein, can inhibit the TGF-β receptor-mediated Smads pathway [16][17].
Hsa_circ_0004018
Li et al. initially studied the expression profile of CircRNA using a microarray, according to their results, hsa_circ_0004018 was significantly down-regulated within the fibrotic mouse liver. In particular, hsa_circ_0004018 down-regulation within HSCs was related to slow fibrosis progression.
Subsequently, an in vitro study found that the hsa_circ_0004018 overexpression suppressed HSCs growth by blocking the cell cycle, as indicated by a higher G0/G1 phase ratio, and the decreased mRNA and protein expression of α-SMA and Col1α1 within HSCs, suggesting that hsa_circ_0004018 may inhibit HSC activation [18]. Further, according to an in vivo study, α-SMA and Col1α1 expression obviously decreases, and liver fibrosis is obviously alleviated following the administration of hsa_circ_0004018-expression lentivirus in mice with CCl4-mediated liver fibrosis [18]. Based on the above findings, telomerase-associated protein 1 (TEP1) is hsa-miR-660-3p’s downstream target. Thus, hsa_circ_0004018 is the potential hsa-miR-660-3p sponge, which can target and suppress TEP1 expression. TEP1 is one of the telomerase components, which assists in telomere maintenance [19], and previous studies have shown that telomeres in hepatocytes or immune cells may be involved in liver fibrosis [20]. In short, the hsa_circ_0004018/hsa-miR-660-3p/TEP1 axis is related to HSC activation and proliferation, which is the potential novel therapeutic strategy for liver fibrosis [18].
Mmu_circ_34116
Zhou et al. used a CircRNA microarray to investigate the differentially expressed CircRNAs using the mouse model of CCl4-mediated liver fibrosis, and discovered 14 CircRNAs with up-regulation and 55 with down-regulation in liver fibrosis tissues [21]. After exploring the expression profiles of mouse fibroblasts JS1 activated by TGF-β1, three CircRNAs were obtained, which displayed consistent expression patterns in both liver fibrotic tissues and JS1 fibroblasts. Among them, mmu_circ_33594 and mmu_circ_35216 expression increased significantly, while mmu_circ_34116 expression considerably declined, indicating that they may be associated with HSC activation [21].
Subsequently, mmu_circ_34116 expression was knocked down after transient mmu_circ_34116 siRNA transfection. According to their results, α-SMA mRNA and protein expression remarkably elevated, indicating that mmu_circ_34116 inhibits HSC activation [21]. Based on further bioinformatics analyses, circRNA_34116 contained miRNA response element (MRE) in miR-22, which showed competitive binding to miR-22, thus exerting indirect regulation on BMP7 target gene transcription [21]. Earlier studies have revealed that treating CCl4-mediated liver fibrosis mice with antisense miR-22 increases BMP7 expression, and remarkably suppresses the development of fibrosis. Therefore, miR-22 possibly enhances liver fibrosis development by suppressing BMP7 [22]. In brief, mmu_circ_34116 suppresses HSCs activation and fibrosis via mmu_circ_34116/miR-22/BMP7 axis [21].
CircDIDO1
CircDIDO1 (ID: hsa_circ_0061137) originates from two to six exons in the DIDO1 gene and is located at the chr20: 61537238-61545758 strand with a 1781-bp genomic length. As a stable circular transcript, CircDIDO1 is initially found to be primarily distributed in the cytoplasm of the human LX-2 cell line. However, Ma et al. observed the low and stable expression of CircDIDO1 within serum exosomes from liver failure cases and predicted that serum exosomal CircDIDO1 showed useful significance in diagnosing liver failure [23]. Thereafter, according to the in vitro research results, CircDIDO1 suppressed DNA synthesis and cell viability, reduced expression of a-SMA and collagen I pro-fibrotic markers, promoted cell apoptosis, and blocked the cell cycle of an LX-2 cell line by blocking the PTEN/AKT pathway [23]. Furthermore, when extracellular CircDIDO1 was added in exosomes obtained from mesenchymal stem cells (MSCs) and later transmitted into HSCs, the results indicated that extracellular CircDIDO1 restrained HSC activation by up-regulating PTEN, and it also inhibited the AKT pathway through sponging miR-141-3p. Mechanically, MSC-originated exosomal CircDIDO1 suppressed HSC activation and liver fibrosis development via miR-141-3p/PTEN/AKT pathway within human liver fibrosis [23], thus shedding new light on developing exogenous CircRNAs to prevent liver fibrosis [23].
CircMTO1
CircMTO1 originates from MTO1 (mitochondrial tRNA translation optimization 1 gene, hsa_circ_0007874) [24], which is recognized as the tumor suppressor of HCC [25]. CircMTO1, which can be detected on chr6:74175931-74176329 (circBase database), has decreased expression within liver tissues from cirrhosis cases, activated HSCs and the CCl4 initiated liver fibrosis model [24]. As suggested by Wang et al., CircMTO1 overexpression inhibited the TGF-β1-mediated HSC activation and cell cycle progression, suppressed α-SMA and type I collagen levels, and relieved liver fibrosis through targeting miR-17-5p and Smad7 [26]. In contrast, based on Zheng et al., miR-181b-5 activated HSC via the PTEN/AKT cascade [27], while Jin et al. further revealed that miR-181b-5p and CircMTO1 were co-distributed within the cytoplasm and interacted with each other. Thereafter, CircMTO1 suppressed HSC activation, which was almost completely restrained via PTEN or miR-181b-5p [24]. Consequently, CircMTO1 inhibited HSC activation through the miR-17-5p/Smad7 axis or miR-181b-5p-mediated PTEN expression, ultimately slowing down liver fibrosis. Such results indicate that CircMTO1 is the potential novel therapeutic target of liver fibrosis. Further, serum CircMTO1 expression is found to evidently decrease among patients with chronic hepatitis B (CHB), which shows negative relation to various stages of fibrosis. Similarly, CircMTO1 down-regulation predicts a dismal prognostic outcome, which is the candidate diagnostic biomarker for CHB patients [26].
2.2. CircRNAs Are Involved in Promoting Liver Fibrosis
Certain CircRNAs show up-regulation within HSCs and liver fibrotic tissues and promote liver fibrosis development via several mechanisms (e.g., playing a role of miRNA sponges, combining with functional miRNAs, and modulating transcription and post-transcription levels of genes) [4][7][28]. The above CircRNAs with up-regulation are the potential diagnostic markers and therapeutic targets of liver fibrosis.
2.2.1. CircRNAs Are Related to Liver Fibrosis by Playing a Role of miRNAs-Binding CeRNAs
CircUbe2k
CircUbe2k (mmu_circ_0001350) is located on chromosome chr5: 65957227-65985787 (465nt), and consists of five exons (exon 2–6) at Ube2k gene locus [7]. CircUbe2k expression is shown to be significantly enhanced in TGF-β1-stimulated LX-2 cells and CCl4-mediated liver fibrosis [7]. However, when CircUbe2k expression decreases by siRNA-circUbe2k transfection within the activated LX-2 cells, α-SMA and Col1α1 mRNA levels subsequently decrease, and LX-2 cell viability and proliferation are strongly suppressed. Further, injection with AAV-siRNA-circUbe2k leads to the inhibition of α-SMA, Col1α1, TGF-β1, and TIMP-1 expression. Specifically, collagen and ECM deposition are reduced in mice with CCl4-mediated liver fibrosis, which thereby suppresses liver fibrosis [7]. From the mechanism perspective, CircUbe2k, as the miRNA sponge, has the capacity to bind to miR-149-5p, so as to regulate the TGF-β2 level. This indicates that it can promote HSCs activation and HF progression via CircUbe2k/miR-149-5p/TGF-β2 axis [7].
CircPWWP2A
The expression of CircPWWP2A (corresponding to hsa_circ_0074837 in humans and mmu_circ_0000254 in mice) significantly increases in LPS- and TGF-β-stimulated HSCs, as well as in liver tissues in mice with liver fibrosis. Further mechanistic studies demonstrate that CircPWWP2A promotes HSC growth and activation as a sponge of miR-203 and miR-223, which then promotes Fstl1 and TLR4 levels, respectively, and eventually promotes liver fibrosis [4].
CircRNA-4099
As suggested by a number of articles, liver fibrosis, usually resulting from oxidative stress [29], represents the eventual common pathway underlying hepatitis of different causes. Hepatitis accounts for one liver dysfunction type that typically indicates various levels of damage to liver cells induced by a variety of pathogenic factors [30].
As an oxidant, H2O2 can significantly increase the cellular ROS levels and α-SMA, type I, and type III collagen production within L02 human hepatocytes, thus inhibiting L02 cell viability dose-dependently and inducing L02 cell fibrosis and injury [31]. The L02 cells are treated with different H2O2 dilutions for mimicking hepatitis in vitro, and the results demonstrate that H2O2 induces severe cell damage and CircRNA-4099 overexpression. Furthermore, the depletion of CircRNA-4099 alleviates the H2O2-induced cell damage, while its overexpression exhibits the opposite effects [31]. Further mechanistic research implies that CircRNA-4099 aggravates H2O2-mediated damage by suppressing miR-706 by activating p38MAPK and keap1/Nrf2 within L02 cells. This suggests that CircRNA-4099 may be a key therapeutic target for hepatitis and fibrosis [31].
CircRNA-0067835
Thymosin β4 (Tβ4) is the 43-amino acid polypeptide with a high conservation degree and the most abundant actin-sequestering protein in cells, which promotes cell growth and survival via the PI3K/AKT signaling pathway [27][32][33]. Zhu et al. applied a circRNA microarray in investigating Tβ4-associated CircRNAs and identified altogether 644 CircRNAs with differential expression in Tβ4-deficient LX-2 cells compared with control cells [28]. Further studies revealed that the CircRNA-0067835 level was remarkably elevated within Tβ4-siRNA LX-2 cells relative to controls, and the CircRNA-0067835 knockout obviously reduced cell growth by inducing G1 arrest while enhancing apoptosis [28]. Moreover, the CircRNA-0067835 level was markedly elevated within the CCl4-treated mouse livers. Finally, CircRNA-0067835 was demonstrated to promote liver fibrosis development by sponging miR-155 for promoting FOXO3a and AKT expression [28].
CircRSF1 and Hsa_circ_0071410
Radioactive liver disease (RILD) has been recognized to be the main complication secondary to radiotherapy for liver cancer [34], which may induce liver failure or even death in some severe cases [35]. The abnormal expression of CircRNA is possibly related to cellular responses to radiation and the biological process of liver fibrosis. Chen et al. elucidated that the Ras-related C3 botulinum toxin substrate 1 (RAC1) and miR-146a-5p levels markedly increased within the human HSCs (LX-2) irradiated with 8 Gy X-rays. RAC1 overexpression remarkably promoted proinflammatory factor production, and α-SMA and collagen I levels, by promoting NF-κB p65 phosphorylation and Bcl-2 expression within irradiated LX-2 cells. However, the overexpression of miR-146a-5p induced via miR-146a-5p mimic transfection repressed RAC1 and Bcl2 levels and decreased the synthesis of α-SMA and type I collagen through inhibiting the phosphorylation of NF-κB p65, JNK1, and Smad1. Therefore, miR-146a-5p reversed the regulation of RAC1 in HSCs [36]. Based on bioinformatics analysis, cricRSF1 (hsa_circ_0023706) contained three MREs of miR-146a-5p [37]. Since CircRSF1 was up-regulated in irradiated LX-2 cells, Chen et al. further co-transfected the CircRSF1 expression vector and miR-146a-5p into LX-2 cells. As a result, CircRSF1 reduced inhibition of miR-146a-5p on the RAC1-triggered activation of NF-κB and JNK/Smad2 pathways, thereby promoting pro-inflammatory factor expression and production, increasing fibrosis marker expression, and finally stimulating the activation of HSCs [36]. Collectively, these results indicate that there is a function regulatory axis involving CircRSF1, miR-146a-5p, and RAC1 within the irradiated HSCs.
Furthermore, as clarified by Chen et al., miR-9-5p expression evidently decreased within the irradiated LX-2 cells, while hsa_circ_0071410 silencing up-regulated miR-9-5p [9]. Meanwhile, hsa_circ_0071410 knockdown significantly reduced α-SMA protein and mRNA expression within the irradiated LX-2, and inhibited the irradiated LX-2 cell growth, probably through inhibiting miR-9-5p. These results indicate that hsa_circ_0071410 promotes irradiated fibrosis via miR-9-5p.
This entry is adapted from the peer-reviewed paper 10.3390/biom13060940
References
- Tao, L.; Ma, W.; Wu, L.; Xu, M.; Yang, Y.; Zhang, W.; Sha, W.; Li, H.; Xu, J.; Feng, R.; et al. Glial cell line-derived neurotrophic factor (GDNF) mediates hepatic stellate cell activation via ALK5/Smad signalling. Gut 2019, 68, 2214–2227.
- Breitkopf-Heinlein, K.; Meyer, C.; Konig, C.; Gaitantzi, H.; Addante, A.; Thomas, M.; Wiercinska, E.; Cai, C.; Li, Q.; Wan, F.; et al. BMP-9 interferes with liver regeneration and promotes liver fibrosis. Gut 2017, 66, 939–954.
- Zhang, C.Y.; Yuan, W.G.; He, P.; Lei, J.H.; Wang, C.X. Liver fibrosis and hepatic stellate cells: Etiology, pathological hallmarks and therapeutic targets. World J. Gastroenterol. 2016, 22, 10512–10522.
- Liu, W.; Feng, R.; Li, X.; Li, D.; Zhai, W. TGF-β- and lipopolysaccharide-induced upregulation of circular RNA PWWP2A promotes hepatic fibrosis via sponging miR-203 and miR-223. Aging 2019, 11, 9569–9580.
- Chen, X.; Li, H.D.; Bu, F.T.; Li, X.F.; Chen, Y.; Zhu, S.; Wang, J.N.; Chen, S.Y.; Sun, Y.Y.; Pan, X.Y.; et al. Circular RNA circFBXW4 suppresses hepatic fibrosis via targeting the miR-18b-3p/FBXW7 axis. Theranostics 2020, 10, 4851–4870.
- Yang, Y.R.; Hu, S.; Bu, F.T.; Li, H.; Huang, C.; Meng, X.M.; Zhang, L.; Lv, X.W.; Li, J. Circular RNA CREBBP Suppresses Hepatic Fibrosis Via Targeting the hsa-miR-1291/LEFTY2 Axis. Front. Pharmacol. 2021, 12, 2515.
- Zhu, S.; Chen, X.; Wang, J.N.; Xu, J.J.; Wang, A.; Li, J.J.; Wu, S.; Wu, Y.Y.; Li, X.F.; Huang, C.; et al. Circular RNA circUbe2k promotes hepatic fibrosis via sponging miR-149-5p/TGF-β2 axis. FASEB J. 2021, 35, e21622.
- Zhou, Y.P.; Lv, X.Y.; Qu, H.; Zhao, K.K.; Fu, L.Y.; Zhu, L.W.; Ye, G.L.; Guo, J.M. Differential expression of circular RNAs in hepatic tissue in a model of liver fibrosis and functional analysis of their target genes. Hepatol. Res. 2019, 49, 324–334.
- Chen, Y.H.; Yuan, B.Y.; Wu, Z.F.; Dong, Y.Y.; Zhang, L.; Zeng, Z.C. Microarray profiling of circular RNAs and the potential regulatory role of hsa_circ_0071410 in the activated human hepatic stellate cell induced by irradiation. Gene 2017, 629, 35–42.
- Zhao, Q.Y.; Liu, J.Y.; Deng, H.; Ma, R.Y.; Liao, J.Y.; Liang, H.X.; Hu, J.X.; Li, J.Q.; Guo, Z.Y.; Cai, J.C.; et al. Targeting Mitochondria-Located circRNA SCAR Alleviates NASH via Reducing mROS Output. Cell 2020, 183, 76–93.e22.
- Petrij, F.; Giles, R.H.; Dauwerse, H.G.; Saris, J.J.; Hennekam, R.C.; Masuno, M.; Tommerup, N.; van Ommen, G.J.; Goodman, R.H.; Peters, D.J.; et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature 1995, 376, 348–351.
- Jia, D.S.; Augert, A.; Kim, D.W.; Eastwood, E.; Wu, N.; Ibrahim, A.H.; Kim, K.B.; Dunn, C.T.; Pillai, S.P.S.; Gazdar, A.F.; et al. Crebbp Loss Drives Small Cell Lung Cancer and Increases Sensitivity to HDAC Inhibition. Cancer Discov. 2018, 8, 1422–1437.
- Menke, L.A.; Gardeitchik, T.; Hammond, P.; Heimdal, K.R.; Houge, G.; Hufnagel, S.B.; Ji, J.; Johansson, S.; Kant, S.G.; Kinning, E.; et al. Further delineation of an entity caused by CREBBP and EP300 mutations but not resembling Rubinstein-Taybi syndrome. Am. J. Med. Genet. A 2018, 176, 862–876.
- Sadeghi, H.; Esmkhani, S.; Pirjani, R.; Amin-Beidokhti, M.; Gholami, M.; Azizi Tabesh, G.; Ghasemi, M.R.; Gachkar, L.; Mirfakhraie, R. CREB-binding protein (CREBBP) and preeclampsia: A new promising target gene. Mol. Biol. Rep. 2021, 48, 2117–2122.
- Bu, F.T.; Zhu, Y.; Chen, X.; Wang, A.; Zhang, Y.F.; You, H.M.; Yang, Y.; Yang, Y.R.; Huang, C.; Li, J. Circular RNA circPSD3 alleviates hepatic fibrogenesis by regulating the miR-92b-3p/Smad7 axis. Mol. Ther. Nucleic Acids 2021, 23, 847–862.
- Ji, D.; Chen, G.F.; Wang, J.C.; Ji, S.H.; Wu, X.W.; Lu, X.J.; Chen, J.L.; Li, J.T. Hsa_circ_0070963 inhibits liver fibrosis via regulation of miR-223-3p and LEMD3. Aging 2020, 12, 1643–1655.
- Hellemans, J.; Preobrazhenska, O.; Willaert, A.; Debeer, P.; Verdonk, P.C.; Costa, T.; Janssens, K.; Menten, B.; Van Roy, N.; Vermeulen, S.J.; et al. Loss-of-function mutations in LEMD3 result in osteopoikilosis, Buschke-Ollendorff syndrome and melorheostosis. Nat. Genet. 2004, 36, 1213–1218.
- Li, S.; Song, F.M.; Lei, X.; Li, J.T.; Li, F.; Tan, H.B. hsa_circ_0004018 suppresses the progression of liver fibrosis through regulating the hsa-miR-660-3p/TEP1 axis. Aging 2020, 12, 11517–11529.
- Harrington, L.; McPhail, T.; Mar, V.; Zhou, W.; Oulton, R.; Bass, M.B.; Arruda, I.; Robinson, M.O. A mammalian telomerase-associated protein. Science 1997, 275, 973–977.
- Barnard, A.; Moch, A.; Saab, S. Relationship between Telomere Maintenance and Liver Disease. Gut Liver 2019, 13, 11–15.
- Zhou, Y.P.; Lv, X.Y.; Qu, H.; Zhao, K.K.; Fu, L.Y.; Zhu, L.W.; Ye, G.L.; Guo, J.M. Preliminary screening and functional analysis of circular RNAs associated with hepatic stellate cell activation. Gene 2018, 677, 317–323.
- Ji, D.; Li, B.; Shao, Q.; Li, F.; Li, Z.B.; Chen, G.F. MiR-22 Suppresses BMP7 in the Development of Cirrhosis. Cell. Physiol. Biochem. 2015, 36, 1026–1036.
- Ma, L.; Wei, J.F.; Zeng, Y.L.; Liu, J.P.; Xiao, E.H.; Kang, Y.H.; Kang, Y. Mesenchymal stem cell-originated exosomal circDIDO1 suppresses hepatic stellate cell activation by miR-141-3p/PTEN/AKT pathway in human liver fibrosis. Drug Deliv. 2022, 29, 440–453.
- Jin, H.; Li, C.X.; Dong, P.H.; Huang, J.T.; Yu, J.L.; Zheng, J.J. Circular RNA cMTO1 Promotes PTEN Expression Through Sponging miR-181b-5p in Liver Fibrosis. Front. Cell Dev. Biol. 2020, 8, 714.
- Han, D.; Li, J.X.; Wang, H.M.; Su, X.P.; Hou, J.; Gu, Y.; Qian, C.; Lin, Y.; Liu, X.; Huang, M.Y.; et al. Circular RNA circMTO1 Acts as the Sponge of MicroRNA-9 to Suppress Hepatocellular Carcinoma Progression. Hepatology 2017, 66, 1151–1164.
- Wang, W.; Dong, R.L.; Guo, Y.; He, J.A.; Shao, C.P.; Yi, P.; Yu, F.J.; Gu, D.Y.; Zheng, J.J. CircMTO1 inhibits liver fibrosis via regulation of miR-17-5p and Smad7. J. Cell. Mol. Med. 2019, 23, 5486–5496.
- Zheng, J.J.; Wu, C.Z.; Xu, Z.Q.; Xia, P.; Dong, P.H.; Chen, B.C.; Yu, F.J. Hepatic stellate cell is activated by microRNA-181b via PTEN/Akt pathway. Mol. Cell. Biochem. 2015, 398, 1–9.
- Zhu, L.; Ren, T.; Zhu, Z.; Cheng, M.; Mou, Q.; Mu, M.; Liu, Y.; Yao, Y.; Cheng, Y.; Zhang, B.; et al. Thymosin-β4 Mediates Hepatic Stellate Cell Activation by Interfering with CircRNA-0067835/miR-155/FoxO3 Signaling Pathway. Cell Physiol. Biochem. 2018, 51, 1389–1398.
- Campana, L.; Iredale, J.P. Regression of Liver Fibrosis. Semin. Liver Dis. 2017, 37, 1–10.
- Lawrence, Y.A.; Dangott, L.J.; Rodrigues-Hoffmann, A.; Steiner, J.M.; Suchodolski, J.S.; Lidbury, J.A. Proteomic analysis of liver tissue from dogs with chronic hepatitis. PLoS ONE 2018, 13, e0208394.
- Li, Y.L.; Gao, X.J.; Wang, Z.H.; Liu, W.; Xu, F.; Hu, Y.J.; Li, Y.N.; Shi, L. Circular RNA 4099 aggravates hydrogen peroxide-induced injury by down-regulating microRNA-706 in L02 cells (Retracted article. See vol. 308, 2022). Life Sci. 2020, 241, 116826.
- Goldstein, A.L.; Hannappel, E.; Kleinman, H.K. Thymosin beta4: Actin-sequestering protein moonlights to repair injured tissues. Trends Mol. Med. 2005, 11, 421–429.
- Adachi, M.; Osawa, Y.; Uchinami, H.; Kitamura, T.; Accili, D.; Brenner, D.A. The forkhead transcription factor FoxO1 regulates proliferation and transdifferentiation of hepatic stellate cells. Gastroenterology 2007, 132, 1434–1446.
- O’Sullivan, B.; Levin, W. Late radiation-related fibrosis: Pathogenesis, manifestations, and current management. Semin. Radiat. Oncol. 2003, 13, 274–289.
- Munoz-Schuffenegger, P.; Ng, S.; Dawson, L.A. Radiation-Induced Liver Toxicity. Semin. Radiat. Oncol. 2017, 27, 350–357.
- Chen, Y.H.; Yuan, B.Y.; Chen, G.W.; Zhang, L.; Zhuang, Y.; Niu, H.; Zeng, Z.C. Circular RNA RSF1 promotes inflammatory and fibrotic phenotypes of irradiated hepatic stellate cell by modulating miR-146a-5p. J. Cell. Physiol. 2020, 235, 8270–8282.
- Ye, W.B.; Lv, Q.; Wong, C.K.A.; Hu, S.; Fu, C.; Hua, Z.; Cai, G.P.; Li, G.X.; Yang, B.B.; Zhang, Y. The Effect of Central Loops in miRNA:MRE Duplexes on the Efficiency of miRNA-Mediated Gene Regulation. PLoS ONE 2008, 3, e1719.
This entry is offline, you can click here to edit this entry!
