Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Colorectal cancer is the second leading cause of cancer-related deaths worldwide. The incidence of this cancer continues to rise, especially in developing countries. Alternative splicing is a normal cellular process that results in the generation of proteins with different structures and functions from a single gene. Colorectal cancer can cause dysregulation of alternative splicing processes to promote its development and growth until it spreads. Dysregulated alternative splicing processes have been shown to promote cancer survival by producing proteins that activate genes known to promote cancer development or deactivate those that inhibit cancer development.
- colorectal cancer
- alternative splicing
- protein arginine methyltransferases
1. Introduction
Colorectal cancer (CRC) has been shown to greatly contribute to mortality, morbidity, and the economic costs of healthcare worldwide. The global burden of the disease is reflected by the reported incidence of the disease being 1.8 million cases, with 0.9 million deaths, and 19 million disability-adjusted life years (DALYs) worldwide [1][2]. According to the GLOBACAN 2020 cancer statistics, CRC is the third- and second-ranked cancer for its overall incidence and mortality worldwide, respectively (Figure 1) [3].
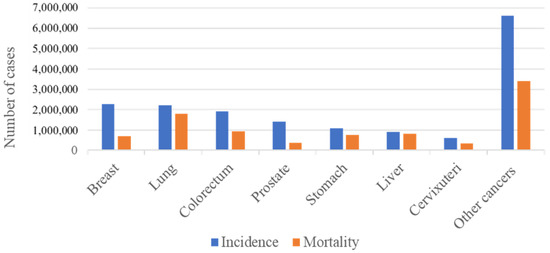
Figure 1. Incidence and mortality numbers for the most prevalent cancers. The figure shows the mortality and incidence numbers of the most prevalent cancers worldwide in the year 2020. Incidence reflects the number of newly diagnosed cases, while mortality reflects the number of deaths related to each cancer. It can be seen that worldwide colorectal cancer is the third most prevalent and accounts for the second most cancer-related deaths.
Furthermore, the incidence of CRC is believed to be on the rise, particularly in low-middle income countries (LMICs) and Sub-Saharan Africa (SSA), which is often associated with socio-economic transitions. Its incidence has been reported to be stabilizing or decreasing in middle-high and high-income populations [1][2][4]. Given the high costs for the screening and treatment of CRC, identifying novel biomarkers for the prediction and therapeutic interventions is urgently needed. Cancer cells arise from the accumulation of several mutations in response to a variety of factors, including epigenetic alterations. Genomic instability promotes the progression from precancerous lesions to carcinoma. The commonly known genomic instability in CRC involves microsatellite instability (MSI), chromosomal instability, and chromosome translocations [1][2]. Cancer cells with genetic alterations have the ability to evade the immune system [3]. These include MSI-high cancers that can avoid recognition by the immune system by undergoing frequent immunoediting resulting in alterations in the major histocompatibility complex (MHC)-antigen presentation pathway [4].
Precursor mRNA (pre-mRNA) splicing is an important post-transcriptional process that occurs in mammalian cells. In this process, introns are removed by an enzyme complex referred to as the spliceosome, and exons are joined back together. This results in a mature mRNA ready for translation into a protein [5]. Several mRNA variants can be formed from a single gene through a process known as alternative splicing (AS). Here, introns are removed and several exons are joined together in different combinations to produce mRNA variants with equal chances to be translated into unique proteins with different, or even opposing, functions [6]. Several studies have reported that about 90–95% of mammalian genes undergo AS and are often associated with cellular homeostasis, differentiation and lineage determination, tissue growth and maintenance, and organ development [5][7]. The genetic and epigenetic alterations in molecules associated with mRNA splicing may result in the generation of aberrant mRNA transcripts which may contribute to tumorigenesis [8].
2. Clinical Significance of AS Events in Cancer
There is growing interest in post-transcriptional splicing factor mutations and their roles in carcinogenesis [9]. Cancers develop as a result of alterations in gene expression and post-transcriptional modifications (PTM). This includes genetic mutations, epigenetic modifications, aberrant alternative splicing (AS), and changes in the transcription of non-coding RNAs such as miRNA. These changes can occur in response to a wide range of factors (environmental and infectious) [10]. In recent years, alternatively spliced events were associated with numerous types of cancer through the development of high throughput technologies. Interestingly, some AS events hold the potential to be explored further and used in the pre-clinical and clinical settings. A study that compared esophageal squamous cell carcinoma (ESCC) tissues to normal tissues found that a total of 45,439 AS events take place in esophageal squamous cells. The study reported that 6019 of the AS events differ significantly in ESCC tissues compared to normal tissues, resulting in differently spliced mRNA and protein isoforms unique to the disease [11]. The study further demonstrated that the splicing factor 3b subunit 4 (SF3B4) was responsible for 102 abnormal AS events in 92 targeted genes. The expression of SF3B4 was associated with survival-related genes in ESCC.
These findings were supported by other studies indicating that heterozygosity for SF3B4 mutations leads to defects in mRNA splicing, particularly exon skipping. Overexpression of SF3B4 in cancer cells also caused mis-splicing of Kruppel-like factor 4 (KLF4), a tumor suppressor-encoding gene, resulting in a non-functional transcript, and therefore promoting carcinogenesis in hepatocellular carcinoma [12][13]. Studies have also identified splicing events specific to CRC. A study by Xiong Y et al., 2018 reported that 34,334 AS events from 8942 genes were identified in CRC tissues. This means that one gene might have almost four AS events on average. Furthermore, the study showed that out of the 34,334 identified AS events, 421 AS events were differentially expressed between samples when they were divided based on clinical features such as age, sex, and OS, as well as tumor size, lymph node status, and metastasis (TNM) stage. However, the differentially expressed AS events were not compared to those expressed in normal tissue.
3. Alternative Splicing (AS) Events in Colorectal Cancer Pathogenesis
The structurally and functionally different proteins that can result from pre-mRNA splicing contribute to genetic diversity in eukaryotic cells [14]. Impaired cellular homeostasis, a major contributor to cancer, is considered to be directly related to aberrant alternatively spliced transcripts. Mutations and changes in the concentration of splice factors may contribute to cancer because alternative splicing governs the production of spliced variants and plays a crucial role in post-transcriptional regulation [15][16]. Through the occurrence of the alternative splicing event such as exon skipping, intron retention, and the choice of alternative splice sites, cancer-specific transcripts, and isoforms are produced which further impact cancer biological processes which include angiogenesis, apoptosis, cell-cycle regulation, metastasis, proliferation and invasion [16][17]. Just as in other types of cancers, AS is a hallmark in the development and progression of CRC. There are aberrant AS events that are reported to be closely associated with CRC progression.
3.1. Implications of lncRNAs AS Events in Colorectal Cancer
Long non-coding RNAs (lncRNAs) have emerged as key regulators in cancer biology, including CRC [18]. To date, a lot of evidence has revealed that long non-coding RNA (lncRNA) molecules are aberrantly expressed in CRC tissues or cells, which regulate gene expression and participate in the occurrence and development of CRC by regulating cell proliferation, cell cycle, epithelial–mesenchymal transition (EMT), drug resistance, and metastasis [19]. Some of the examples of lncRNAs that are aberrantly expressed and play a pivotal role in CRC carcinogenesis includes LncRNA-SNHG11, LncRNA-RPPH1, LINC01106, lncRNA-APC1, and lncRNA-AK028845 [19]. LncRNAs are also known to directly affect the function of micro-RNAs (miRNAs). Micro-RNAs (miRNAs) are part of the non-coding RNA (ncRNA) family, and miRNAs are smaller transcripts that are 18–22 base pairs long [20]. miRNAs are one of the small molecules known to regulate biological processes via the splicing of mRNA to generate alternate transcripts. For example, alternative transcripts such as miR-583-3p and miR-1273-3p were previously associated with cell growth and proliferation in colon cancers [21]. High expression of miR-340-5b is reported to promote invasion, metabolism, and EMT in CRC through the activation of the ERK signaling pathway [18][21]. There is a significant increase in the number of novel lncRNAs associated with CRC. LINC00662 is a lncRNA which plays a crucial role in colon cancer progression through the activation of the ERK signaling pathway [18][19]. Another lncRNA of interest is the colon cancer-associated transcript 1 (CCAT1), which was discovered by Nissan et al. CCAT1 has been shown to be overexpressed in various cancer types, including CRC [22]. Recent research suggests that CCAT1 promotes colon cancer cell growth by increasing expression of the oncoprotein c-MYC and the oncogenic mRNA tumor suppressor candidate 3 (TUSC3), the target of miR-181b-5p in CRC cells, thus increasing glucose metabolism to fuel colon cancer cell growth. This promotes colon cancer cell migration and invasion by accelerating the EMT process and negatively modulating miR-218 and hsa-miR-4679; and suppresses apoptosis [19]. Several other miRNAs are reported to be dysregulated in colon cancers, including but not limited to hsa-miR-585-3p, hsa-miR-1273, hsa-miR-340-5p, hsa-miR-374b-5p, and hsa-miR-335-5p [23]. Understanding the intricate network of alternative splicing in lncRNAs and its impact on CRC pathogenesis holds great promise for the development of novel diagnostic and therapeutic approaches. Targeting specific alternatively spliced lncRNA isoforms or modulating splicing factors could offer potential strategies for precision medicine in colorectal cancer.
3.2. Other AS Variants Associated with Colorectal Cancer
As mentioned above, AS does not occur independently. Instead, this procedure is linked to other cellular mechanisms that are frequently manipulated during carcinogenesis, such as apoptosis, chemoresistance, angiogenesis, metastasis, cell-cycle progression, proliferation, and invasion. For instance, the biological process of apoptosis relies on a delicate equilibrium between pro- and anti-apoptotic factors to determine the fate of cells [8]. Intriguingly, it has been shown that AS generates opposing regulators of apoptosis, suggesting that AS plays a critical part in a cell’s life-or-death decision-making [22]. BCL2 Like 1 (BCL2L1) is one of the many regulators of apoptosis and a member of the BCL2 Apoptosis Regulator (BCL2) family [8][24]. Alternative splicing of BCL2L1 (BCL-X) results in either a long anti-apoptotic variant (BCL-xl) or a short pro-apoptotic variant (BCL-xs) and this splicing switch is facilitated by the SRSF1 splicing factor [25][26]. High expression of SRSF1 was reported to generate two isoforms of MAPK interacting serine/threonine kinase 2 (MNK2), namely MNK2a and MNK2b, in CRC cells [27].
The high expression of SRSF1 causes an imbalance between the two isoforms with an upregulation of MNK2b and downregulation of MNK2a. Consequently, this inhibits the p38a-MAPK signaling pathway, which results in increased cell proliferation and a decreased rate of apoptosis [27]. Although high expression of the variants contributing to the imbalance in apoptotic factors is reported to play a role in the development and progression of different types of cancers, the specific mechanisms are not yet fully understood [27][28][29].
4. Contribution of PRMTs and SFKs Regulatory Networks in CRC Carcinogenesis
4.1. The Role of Alternatively Spliced Transcripts of PRMTs in Colorectal Cancer
Regardless of the current knowledge regarding the contribution of AS events/variants to the development of cancers, the contribution of AS in CRC remains understudied. This is particularly true when it comes to identifying AS events in splicing regulators such as PRMTs and SFKs and their contributions to CRC. PRMTs are a group of enzymes that catalyze arginine methylation, which is currently recognized as a widespread post-transcriptional modification in many proteins [30]. The pivotal role played by arginine methylation in mammals is well recognized and includes, but is not limited to, splicing regulation, RNA metabolism, DNA damage repair, phase separation, and signal transduction [31]. There are three different classes of arginine methyltransferases (PRMT I, PRMT II, and PRMT III) that have been identified based on the final end product (when the methyl group is bonded to the R residue. The formation of monomethylarginine (MMA) is the initial product for all classes of PRMTs [32]. The subsequent methylation process varies between enzyme classes. PRMTs 1, 2, 3, 4, 6, and 8 are class I arginine methyltransferases, which catalyze the conversion of MMA into asymmetric demethylated arginine (ADMA) [32][33]. Unlike type I PRMTs, PRMT5 and PRMT9 are class II arginine methyltransferases, which further catalyze MMA conversion into symmetrically dimethylated arginine (SDMA), whilst PRMT7 is the only enzyme in the group of class III arginine methyltransferases, and functions to catalyze the production of MMA [32][33].
Since PRMTs play an important role in arginine methylation, PRMTs are involved in the same processes that require arginine methylation, including the transcriptional and post-transcriptional regulation of gene expression, DNA damage repair, cell-cycle check-points, mRNA processing and translation, as well as intracellular signaling during development and disease progression, particularly in cancers [9][32][33][34]. Over the past decades, studies have shown dysregulation of PRMTs to be associated with cancer progression and metastasis in mammals but the full scope on how the alternatively spliced PRMTs (PRMT isoforms) play a role in tumorigenesis is not yet clearly understood. A study by Adamopoulos et al., 2019 identified a number of AS events that resulted in multiple PRMT1 transcripts which are predicted to encode new protein isoforms [35]. Amongst the pool of PRMT1 variants, two splice variants of PRMT1 (variants v.1 and v.2) were reported to be significantly upregulated in CRC and their overexpression was associated with the nodal status and histological grade of tumors in colon cancer [35][36][37][38].
The AS event PRMT1-51042-ES, reported to be highly expressed by cytotoxic T-helper cells, was identified as an independent predictor of overall survival, genomic instability, and poor prognosis in CRC [39]. PRMT1∆arm, a variant of PRMT1, is missing exons crucial for organizing the dimerization domain necessary for enzymatic activity. As a result, PRMT1∆arm is unable to methylate arginines, but retains the chromatin-binding capacity, competitively limiting the binding of active PRMT1 and ultimately leading to increased chances of malignancy [40]. Given these findings, PRMT1 variants and the AS events leading to these variants may serve as useful prognostic, diagnostic, and/or therapeutic biomarkers for CRC. However, further studies are needed to determine if other types of PRMTs may elicit the same effect in CRCs.
Alterations in splicing factor expression appear to be a significant cause of aberrant splicing profiles, although the processes behind this shift in splicing factor expression in tumors remain poorly understood. Apart from PRMTs, a group of enzymes known as splicing factor kinases (SFKs), which play a role in AS, have been investigated [40]. Serine/arginine protein kinase 1 (SRPK1) is reported to play an important part in AS regulation through phosphorylation of different splicing factors rich in serine/arginine domains (SR proteins), including serine/arginine rich splicing factor 1 (SRSF1) [41][42][43]. Similar to PRMTs, SRPK1 is reported to be overexpressed in many types of malignancies, including CRC. The expression levels of SRPK1 were associated with clinical factors such as TNM staging, and poor disease prognosis or outcome [44][45][46][47][48]. The proper regulation of SRPK1 is crucial in the maintenance of normal physiologic and pathological states in eukaryotic cells, including splice site selection, mRNA export, spliceosome assembly, and translation [49].
4.2. The Role of VEGF in CRC and Metastasis
Vascular endothelial growth factor (VEGF) is a multifunctional cytokine that is involved in angiogenesis through the binding and activation of receptors (VEGFR 1 and 2) on endothelial cells [50][51]. VEGF can undergo alternative splicing to form various isoforms. In particular two of these isoforms, VEGF165b and VEGF165 are formed via the selection of the proximal splice site (SPP) and distal splice site (DSS) in the terminal of exon 8 [50].
Dysregulation of SRPK1 is believed to play a role in the splicing switch from the VEGF165b to the VEGF165 isoform. VEGF165 has been shown to promote cell growth and migration [52]. SRPK1 facilitates the splicing switch of VEGF165b to the VEGF165 isoform by phosphorylating the splicing factor (SRSF1) and promoting proximal splice site usage, ultimately leading to the increased expression of the proangiogenic VEGF165 isoform [51]. Furthermore, the dysregulated expression of SRPK1 in breast cancer increases the phosphorylation of RNA-binding motif protein 4 (RBM4). This leads to the production of RBM4-specific splicing variants of myeloid cell leukemia 1 (MCL-1) and Insulin receptor (IR). The isoforms MCL-1s and IR-B lead to decreased or inhibited apoptosis of malignant cells [53]. Although there is limited knowledge of the specific AS events/variants of SRPK1, the current evidence is clear that dysregulation of SRPK1 affects the phosphorylation of splicing factors, eventually contributing to angiogenesis and tumorigenesis. Hence the available literature suggests that SRPK1 has the potential to function as either an oncogene or tumor-suppressor gene.
5. Therapeutic Potential of Splicing Disrupter Drugs in Cancer and Colorectal Cancer
Large-scale genomic studies concentrating on single-cell RNA sequencing and characterization have proven to be powerful methods to establish how protein-coding and non-coding RNA transcription and processing are dysregulated in numerous malignancies, thus providing insight into the variety and complexity of tumors [54]. In recent years, substantial evidence gathered thus far suggests that the identification of cancer-specific AS variations has the potential to provide novel therapeutic targets in cancer patients [34][54][55]. Different therapeutic strategies have been used to target the complex mechanisms of AS, while another focus has been on the use of whole transcriptome sequencing to identify novel therapeutic targets. Some therapeutic strategies include targeting trans-regulatory factors of splicing, including the spliceosome complex and splicing regulating factors. Another strategy involves the use of splice-switching oligonucleotides (SSOs), which have been used to correct aberrant AS or induce the expression of a splice variant, and another option is the targeting of a novel CRC-relevant splice variant for therapeutic purposes [55][56][57].
Post-transcriptional modification remains the most important process associated with spliceosome functions, and efforts have been made in developing splicing disrupter drugs/inhibitors that specifically target PTM [54]. Dysregulation of PTM can alter the function of splicing factors and several compounds that have the ability to inhibit different modifications, altering PTM. These include inhibitors of CLKs (CDC-like kinases), SRPKs, and PRMTs, which have been screened, with some showing promise as anti-cancer drugs [57]. One important PTM is methylation by PRMT5, a type II PRMT which is critical in the recruitment and assembly of spliceosome components [16]. The inhibition of PRMT5 as well as PRMT1 and CARM1 have been shown to cause splicing inhibition and display anti-cancer properties in several cancers [58].
A vast number of patents have been filed for PRMT inhibitors from both academic laboratories and the pharmaceutical industry [59]. JNJ-64619178 and GSK3326595 are some examples of PRMT5 inhibitors that are reported to be in human phase-1 clinical trials in patients with advanced or recurrent solid tumors [60]. The treatment of THP-1 cells, a leukemia monocytic cell line with a specific PRMT5 inhibitor (EPZ015666), was reported to decrease levels of SDMA methylation and affect cell proliferation negatively [58]. It is worth noting that these inhibitors can also be used in combination with other drugs/inhibitors. For example, GSK3326595 was used together with anti-PD1 therapy in hepatocellular carcinoma (HCC) and improved efficacy was noticed, suggesting that this combination might be worth testing in future HCC clinical trials [61]. However, the presence of multiple PRMT isoforms with distinct functions can cause interpretation to become difficult, as such PRMT inhibitors must exhibit isoform specificity, being able to target one isoform enzyme. Non-specific global inhibitors of methyltransferase cannot be used to precisely target isoforms or be used in studies to elucidate the function of various isoforms. The desirable solution to this is the development of a potent and isoform-selective small-molecule inhibitor.
PRMT1, -3, -4, -6, -7, and -8 possess a region known as cavity-2, beneath the dimerization arm. This region is responsible for dimerization and the activity of these PRMTs [62]. The sequences of amino acids lining the cavity differ amongst different PRMTs as well as different isoforms. The differences of the residue sequences amongst different isoforms may allow for the specific targeting of different isoforms [63]. Isoforms also complicate the assessment of the efficiency of inhibitors. This is normally assessed through IC 50 values or the inhibition constant K [64]. An example of one class of these small inhibitors is the ethanediamine-heterocycle compounds. These compounds appear to selectively inhibit different PRMTs and different PRMT isoforms, with different members of this class being able to act as a pan-PRMT inhibitor or be selective for only one isoform [65].
In patients with melanoma, GSK3326595 in combination with Palbociclib (CDK4/6) inhibitor may assist in decreasing the chances of drug resistance [16]. In contrast to PRMT5, PRMT1 catalyzes ADMA and is overexpressed in multiple cancers [16][17]. Overexpression of PRMT1 reduces the expression of RBM15, which is reported to play a role in hematopoiesis and subsequently affects megakaryocyte terminal differentiation [60]. A completed phase I clinical trial with PRMT1 (GSK3368715) inhibitor was reported to inhibit cancer cell growth in patients with advanced solid tumors and diffuse large B cell lymphoma (DLBCL) [54]. A pre-clinical study using a combination of inhibitors of PRMT1 (MS023) and PRMT5 (EPZ015666) demonstrated an efficient anti-cancer effect in lung cancer and pancreatic cancer cell lines [66]. Currently, there are two CLK (SM08502 and CTX-712) inhibitors that are available for oral consumption which were reported to be in phase I clinical trials in the year 2020 [67]. These inhibitors are reported to target the phosphorylation process of SRSF6 and enlarge nuclear speckle, and have shown great potential to move to phase II clinical trials [67]. Other inhibitors showing great potential are two SRPK (NCT04247256 and NCT04652206) inhibitors, which are reported to be in phase II clinical trials [67]. These inhibitors are administered together with docetaxel and have shown anti-tumour activities in triple-negative breast cancer cells. Currently, there are limited reports on inhibitors in clinical trials that are specific to CRC. Given the data on targeting cancer through PTM inhibition, there is limited data on using splicing disrupter drugs/inhibitors in colorectal cancer. Exploiting the alternative splicing machinery may help in understanding the downstream pathways regulated by PRMTs and splicing factors. This may lead to the discovery of novel opportunities that can be used to exploit the vulnerability of colorectal cancer to splicing inhibitors. These new therapeutic strategies offer great potential to treat CRC, most particularly in low- and middle-income countries.
This entry is adapted from the peer-reviewed paper 10.3390/cancers15153999
References
- Awedew, A.F.; Asefa, Z.; Belay, W.B. Burden and trend of colorectal cancer in 54 countries of Africa 2010–2019: A systematic examination for Global Burden of Disease. BMC Gastroenterol. 2022, 22, 204.
- Kwakye, G.; Dally, C.K. Colorectal cancer screening in sub-Saharan Africa. Lancet Glob. Health 2022, 10, e938–e939.
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249.
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017, 66, 683–691.
- Wang, E.; Aifantis, I. RNA Splicing and Cancer. Trends Cancer 2020, 6, 631–644.
- Öther-Gee Pohl, S.; Myant, K.B. Alternative RNA splicing in tumour heterogeneity, plasticity and therapy. Dis. Model. Mech. 2022, 15, dmm049233.
- Baralle, F.E.; Giudice, J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 2017, 18, 437–451.
- Sciarrillo, R.; Wojtuszkiewicz, A.; Assaraf, Y.G.; Jansen, G.; Kaspers, G.J.; Giovannetti, E.; Cloos, J. The role of alternative splicing in cancer: From oncogenesis to drug resistance. Drug Resist. Updat. 2020, 53, 100728.
- Ouyang, J.; Zhang, Y.; Xiong, F.; Zhang, S.; Gong, Z.; Yan, Q.; He, Y.; Wei, F.; Zhang, W.; Zhou, M.; et al. The role of alternative splicing in human cancer progression. Am. J. Cancer Res. 2021, 11, 4642–4667.
- Iwai, K.; Yaguchi, M.; Nishimura, K.; Yamamoto, Y.; Tamura, T.; Nakata, D.; Dairiki, R.; Kawakita, Y.; Mizojiri, R.; Ito, Y.; et al. Anti-tumor efficacy of a novel CLK inhibitor via targeting RNA splicing and MYC-dependent vulnerability. EMBO Mol. Med. 2018, 10, e8289.
- Ding, J.; Li, C.; Cheng, Y.; Du, Z.; Wang, Q.; Tang, Z.; Song, C.; Xia, Q.; Bai, W.; Lin, L.; et al. Alterations of RNA splicing patterns in esophagus squamous cell carcinoma. Cell Biosci. 2021, 11, 36.
- Marques, F.; Tenney, J.; Duran, I.; Martin, J.; Nevarez, L.; Pogue, R.; Krakow, D.; Cohn, D.H.; Li, B. Altered mRNA Splicing, Chondrocyte Gene Expression and Abnormal Skeletal Development due to SF3B4 Mutations in Rodriguez Acrofacial Dysostosis. PLoS Genet. 2016, 12, e1006307.
- Shen, Q.; Eun, J.W.; Lee, K.; Kim, H.S.; Yang, H.D.; Kim, S.Y.; Lee, E.K.; Kim, T.; Kang, K.; Kim, S.; et al. Barrier to autointegration factor 1, procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3, and splicing factor 3b subunit 4 as early-stage cancer decision markers and drivers of hepatocellular carcinoma. Hepatology 2018, 67, 1360–1377.
- Warzecha, C.C.; Carstens, R.P. Complex changes in alternative pre-mRNA splicing play a central role in the epithelial-to-mesenchymal transition (EMT). Semin. Cancer Biol. 2012, 22, 417–427.
- Liu, Q.; Fang, L.; Wu, C. Alternative Splicing and Isoforms: From Mechanisms to Diseases. Genes 2022, 13, 401.
- Chen, Y.; Huang, M.; Liu, X.; Huang, Y.; Liu, C.; Zhu, J.; Fu, G.; Lei, Z.; Chu, X. Alternative splicing of mRNA in colorectal cancer: New strategies for tumor diagnosis and treatment. Cell Death Dis. 2021, 12, 752.
- Pal, S.; Gupta, R.; Davuluri, R.V. Alternative transcription and alternative splicing in cancer. Pharmacol. Ther. 2012, 136, 283–294.
- Sun, X.; Lin, F.; Sun, W.; Zhu, W.; Fang, D.; Luo, L.; Li, S.; Zhang, W.; Jiang, L. Exosome-transmitted miRNA-335-5p promotes colorectal cancer invasion and metastasis by facilitating EMT via targeting RASA1. Mol. Ther. Nucleic Acids 2021, 24, 164–174.
- Cheng, B.; Rong, A.; Zhou, Q.; Li, W. LncRNA LINC00662 promotes colon cancer tumor growth and metastasis by competitively binding with miR-340-5p to regulate CLDN8/IL22 co-expression and activating ERK signaling pathway. J. Exp. Clin. Cancer Res. 2020, 39, 5.
- Liu, C.; Yang, J.; Wu, H.; Li, J. Downregulated miR-585-3p promotes cell growth and proliferation in colon cancer by upregulating PSME3. OncoTargets Ther. 2019, 12, 6525–6534.
- Peng, S.; Luo, Y.; Chen, L.; Dai, K.; Wang, Q. lncRNAELFN1-AS1 enhances the progression of colon cancer by targeting miR-4270 to upregulate, AURKB. Open Med. 2022, 17, 1999–2012.
- Liau, X.L.; Salvamani, S.; Gunasekaran, B.; Chellappan, D.K.; Rhodes, A.; Ulaganathan, V.; Tiong, Y.L. CCAT 1-A Pivotal Oncogenic Long Non-Coding RNA in Colorectal Cancer. Br. J. Biomed. Sci. 2023, 80, 11103.
- Kim, W.R.; Park, E.G.; Lee, Y.J.; Bae, W.H.; Lee, D.H.; Kim, H.S. Integration of TE Induces Cancer Specific Alternative Splicing Events. Int. J. Mol. Sci. 2022, 23, 10918.
- Singh, R.; Letai, A.; Sarosiek, K. Regulation of apoptosis in health and disease: The balancing act of BCL-2 family proteins. Nat. Rev. Mol. Cell Biol. 2019, 20, 175–193.
- Stevens, M.; Oltean, S. Modulation of the apoptosis gene Bcl-x function through alternative splicing. Front. Genet. 2019, 10, 804.
- Makhafola, T.J.; Mbele, M.; Yacqub-Usman, K.; Hendren, A.; Haigh, D.B.; Blackley, Z.; Meyer, M.; Mongan, N.P.; Bates, D.O.; Dlamini, Z. Apoptosis in Cancer Cells Is Induced by Alternative Splicing of hnRNPA2/B1 Through Splicing of Bcl-x, a Mechanism that Can Be Stimulated by an Extract of the South African Medicinal Plant, Cotyledon orbiculata. Front. Oncol. 2020, 10, 547392.
- Maimon, A.; Mogilevsky, M.; Shilo, A.; Golan-Gerstl, R.; Obiedat, A.; Ben-Hur, V.; Lebenthal-Loinger, I.; Stein, I.; Reich, R.; Beenstock, J.; et al. Mnk2 alternative splicing modulates the p38-MAPK pathway and impacts Ras-induced transformation. Cell Rep. 2014, 7, 501–513.
- De Necochea-Campion, R.; Shouse, G.P.; Zhou, Q.; Mirshahidi, S.; Chen, C.S. Aberrant splicing and drug resistance in AML. J. Hematol. Oncol. 2016, 9, 85.
- Wang, B.D.; Lee, N.H. Aberrant RNA Splicing in Cancer and Drug Resistance. Cancers 2018, 10, 458.
- Wei, H.H.; Fan, X.J.; Hu, Y.; Tian, X.X.; Guo, M.; Mao, M.W.; Fang, Z.Y.; Wu, P.; Gao, S.X.; Peng, C.; et al. A systematic survey of PRMT interactomes reveals the key roles of arginine methylation in the global control of RNA splicing and translation. Sci. Bull. 2021, 66, 1342–1357.
- Blanc, R.S.; Richard, S. Arginine Methylation: The Coming of Age. Mol. Cell 2017, 65, 8–24.
- Basera, A.; Hull, R.; Demetriou, D.; Bates, D.O.; Kaufmann, A.M.; Dlamini, Z.; Marima, R. Competing Endogenous RNA (ceRNA) Networks and Splicing Switches in Cervical Cancer: HPV Oncogenesis, Clinical Significance and Therapeutic Opportunities. Microorganisms 2022, 10, 1852.
- Li, W.J.; He, Y.H.; Yang, J.J.; Hu, G.S.; Lin, Y.A.; Ran, T.; Peng, B.L.; Xie, B.L.; Huang, M.F.; Gao, X.; et al. Profiling PRMT methylome reveals roles of hnRNPA1 arginine methylation in RNA splicing and cell growth. Nat. Commun. 2021, 12, 1946.
- Bessa, C.; Matos, P.; Jordan, P.; Gonçalves, V. Alternative Splicing: Expanding the Landscape of Cancer Biomarkers and Therapeutics. Int. J. Mol. Sci. 2020, 21, 9032.
- Adamopoulos, P.G.; Mavrogiannis, A.V.; Kontos, C.K.; Scorilas, A. Novel alternative splice variants of the human protein arginine methyltransferase 1 (PRMT1) gene, discovered using next-generation sequencing. Gene 2019, 699, 135–144.
- Mathioudaki, K.; Papadokostopoulou, A.; Scorilas, A.; Xynopoulos, D.; Agnanti, N.; Talieri, M. The PRMT1 gene expression pattern in colon cancer. Br. J. Cancer 2008, 99, 2094–2099.
- Zou, J.; Shen, W.; Zhang, Y.; Ying, S. The Role of Protein Arginine Methyltransferase 1 in Gastrointestinal Cancers; IntechOpen: London, UK, 2021.
- Thiebaut, C.; Eve, L.; Poulard, C.; Le Romancer, M. Structure, Activity, and Function of PRMT1. Life 2021, 11, 1147.
- Ding, Q.; Hou, Z.; Zhao, Z.; Chen, Y.; Zhao, L.; Xiang, Y. Identification of the prognostic signature based on genomic instability-related alternative splicing in colorectal cancer and its regulatory network. Front. Bioeng. Biotechnol. 2022, 10, 841034.
- Zheng, M.; Niu, Y.; Bu, J.; Liang, S.; Zhang, Z.; Liu, J.; Guo, L.; Zhang, Z.; Wang, Q. ESRP1 regulates alternative splicing of CARM1 to sensitize small cell lung cancer cells to chemotherapy by inhibiting TGF-β/Smad signaling. Aging 2021, 13, 3554–3572.
- Giannakouros, T.; Nikolakaki, E.; Mylonis, I.; Georgatsou, E. Serine-arginine protein kinases: A small protein kinase family with a large cellular presence. FEBS J. 2011, 278, 570–586.
- Van Roosmalen, W.; Le Dévédec, S.E.; Golani, O.; Smid, M.; Pulyakhina, I.; Timmermans, A.M.; Look, M.P.; Zi, D.; Pont, C.; De Graauw, M.; et al. Tumor cell migration screen identifies SRPK1 as breast cancer metastasis determinant. J. Clin. Investig. 2015, 125, 1648–1664.
- Li, X.H.; Song, J.W.; Liu, J.L.; Wu, S.; Wang, L.S.; Gong, L.Y.; Lin, X. Serine-arginine protein kinase 1 is associated with breast cancer progression and poor patient survival. Med. Oncol. 2014, 31, 83.
- Malvi, P.; Janostiak, R.; Chava, S.; Manrai, P.; Yoon, E.; Singh, K.; Harigopal, M.; Gupta, R.; Wajapeyee, N. LIMK2 promotes the metastatic progression of triple-negative breast cancer by activating SRPK1. Oncogenesis 2020, 9, 77.
- Yi, N.; Xiao, M.; Jiang, F.; Liu, Z.; Ni, W.; Lu, C.; Ni, R.; Chen, W. SRPK1 is a poor prognostic indicator and a novel potential therapeutic target for human colorectal cancer. OncoTargets Ther. 2018, 11, 5359–5370.
- Liu, H.; Gong, Z.; Li, K.; Zhang, Q.; Xu, Z.; Xu, Y. SRPK1/2 and PP1α exert opposite functions by modulating SRSF1-guided MKNK2 alternative splicing in colon adenocarcinoma. J. Exp. Clin. Cancer Res. 2021, 40, 75.
- Duggan, W.P.; O’Connell, E.; Prehn, J.H.; Burke, J.P. Serine-Arginine Protein Kinase 1 (SRPK1): A systematic review of its multimodal role in oncogenesis. Mol. Cell. Biochem. 2022, 477, 2451–2467.
- Li, Q.; Zeng, C.; Liu, H.; Yung, K.W.Y.; Chen, C.; Xie, Q.; Zhang, Y.; Wan, S.W.C.; Mak, B.S.W.; Xia, J.; et al. Protein-Protein Interaction Inhibitor of SRPKs Alters the Splicing Isoforms of VEGF and Inhibits Angiogenesis. iScience 2021, 24, 102423.
- Bates, D.O.; Mavrou, A.; Qiu, Y.; Carter, J.G.; Hamdollah-Zadeh, M.; Barratt, S.; Gammons, M.V.; Millar, A.B.; Salmon, A.H.; Oltean, S.; et al. Detection of VEGF-A(xxx)b isoforms in human tissues. PLoS ONE 2013, 8, e68399.
- Bates, D.O.; Cui, T.G.; Doughty, J.M.; Winkler, M.; Sugiono, M.; Shields, J.D.; Peat, D.; Gillatt, D.; Harper, S.J. VEGF165b, an inhibitory splice variant of vascular endothelial growth factor, is down-regulated in renal cell carcinoma. Cancer Res. 2002, 62, 4123–4131.
- Li, X.; Kim, W.; Arif, M.; Gao, C.; Hober, A.; Kotol, D.; Strandberg, L.; Forsström, B.; Sivertsson, Å.; Oksvold, P.; et al. Discovery of Functional Alternatively Spliced PKM Transcripts in Human Cancers. Cancers 2021, 13, 348.
- Varey, A.H.R.; Rennel, E.S.; Qiu, Y.; Bevan, H.S.; Perrin, R.M.; Raffy, S.; Dixon, A.R.; Paraskeva, C.; Zaccheo, O.; Hassan, A.B.; et al. VEGF165b, an antiangiogenic VEGF-A isoform, binds and inhibits bevacizumab treatment in experimental colorectal carcinoma: Balance of pro- and antiangiogenic VEGF-A isoforms has implications for therapy. Br. J. Cancer 2008, 98, 1366–1379.
- Stevens, M.; Oltean, S. Modulation of Receptor Tyrosine Kinase Activity through Alternative Splicing of Ligands and Receptors in the VEGF-A/VEGFR Axis. Cells 2019, 8, 288.
- Urbanski, L.M.; Leclair, N.; Anczuków, O. Alternative-splicing defects in cancer: Splicing regulators and their downstream targets, guiding the way to novel cancer therapeutics. Wiley Interdiscip. Rev. RNA 2018, 9, e1476.
- Mayer, S.; Hirschfeld, M.; Jaeger, M.; Pies, S.; Iborra, S.; Erbes, T.; Stickeler, E. RON alternative splicing regulation in primary ovarian cancer. Oncol. Rep. 2015, 34, 423–430.
- Sahin, I.; George, A.; Seyhan, A.A. Therapeutic Targeting of Alternative RNA Splicing in Gastrointestinal Malignancies and Other Cancers. Int. J. Mol. Sci. 2021, 22, 11790.
- Bonnal, S.C.; López-Oreja, I.; Valcárcel, J. Roles and mechanisms of alternative splicing in cancer—Implications for care. Nat. Rev. Clin. Oncol. 2020, 17, 457–474.
- Desterro, J.; Bak-Gordon, P.; Carmo-Fonseca, M. Targeting mRNA processing as an anticancer strategy. Nat. Rev. Clin. Oncol. 2020, 19, 112–129.
- Medina-Franco, J. Epi-Informatics: Discovery and Development of Small Molecule Epigenetic Drugs and Probes; Elsevier Science: Amsterdam, The Netherlands, 2016.
- Rahman, M.A.; Nasrin, F.; Bhattacharjee, S.; Nandi, S. Hallmarks of Splicing Defects in Cancer: Clinical Applications in the Era of Personalized Medicine. Cancers 2020, 12, 1381.
- Radzisheuskaya, A.; Shliaha, P.V.; Grinev, V.; Lorenzini, E.; Kovalchuk, S.; Shlyueva, D.; Gorshkov, V.; Hendrickson, R.C.; Jensen, O.N.; Helin, K. PRMT5 methylome profiling uncovers a direct link to splicing regulation in acute myeloid leukemia. Nat. Struct. Mol. Biol. 2019, 26, 999–1012.
- Zhou, R.; Xie, Y.; Hu, H.; Hu, G.; Patel, V.S.; Zhang, J.; Yu, K.; Huang, Y.; Jiang, H.; Liang, Z.; et al. Molecular mechanism underlying PRMT1 dimerization for SAM binding and methylase activity. J. Chem. Inf. Model. 2015, 55, 2623–2632.
- Antonysamy, S.; Bonday, Z.; Campbell, R.M.; Doyle, B.; Druzina, Z.; Gheyi, T.; Han, B.; Jungheim, L.N.; Qian, Y.; Rauch, C.; et al. Crystal structure of the human PRMT5: MEP50 complex. Proc. Natl. Acad. Sci. USA 2012, 109, 17960–17965.
- Copeland, R.A. Evaluation of Enzyme Inhibitors in Drug Discovery: A Guide for Medicinal Chemists and Pharmacologists; John Wiley & Sons: New York, NY, USA, 2013.
- Di Francesco, M.E.; Jones, P.; Mcafoos, T.J. Ethanediamine-Heterocycle Derivatives as Inhibitors of Protein Arginine Methyltransferases. U.S. Patent 11,028,083, 8 June 2021.
- Siu, L.L.; Rasco, D.W.; Vinay, S.P.; Romano, P.M.; Menis, J.; Opdam, F.L.; Heinhuis, K.M.; Egger, J.L.; Gorman, S.A.; Parasrampuria, R. METEOR-1: A phase I study of GSK3326595, a first-in-class protein arginine methyltransferase 5 (PRMT5) inhibitor, in advanced solid tumours. Ann. Oncol. 2019, 30, v159.
- Araki, S.; Ohori, M.; Yugami, M. Targeting pre-mRNA splicing in cancers: Roles, inhibitors, and therapeutic opportunities. Front. Oncol. 2023, 13, 1152087.
This entry is offline, you can click here to edit this entry!
