Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Global climate change and population growth are persistently posing threats to natural resources (e.g., freshwater) and agricultural production. Crassulacean acid metabolism (CAM) evolved from C3 photosynthesis as an adaptive form of photosynthesis in hot and arid regions. It features the nocturnal opening of stomata for CO2 assimilation, diurnal closure of stomata for water conservation, and high water-use efficiency.
- Crassulacean acid metabolism
- C3 to CAM transition
- facultative CAM
1. Introduction
Drastic climate change over the past decades can be reflected by the alternation in atmospheric CO2 levels, tropospheric ozone concentrations, and other environmental indicators [1]. Climate change is not only affecting ecosystems, but also agriculture, food production, land, and water resources [2]. Arid or semi-arid land accounts for around 41% of the total surface on Earth, and it is expanding [3]. In 2035, global desertification is projected to be 65% of the total land surface in the subtropical regions [4]. With the rapid growth of the human population, the demand for food is increasing, and it is anticipated to surge by 70%. The current rate of global crop productivity only increases by ~2% per year, which cannot meet the demand for food [5]. To worsen the situation, the global decrease in freshwater from 1980 to 2015 has caused a 20.6% and 39.3% yield reduction in wheat and maize, respectively [6].
Photosynthesis, a pivotal biological process essential to all life, provides food and most of the energy resources [7]. There are three major modes of photosynthesis in vascular plants to assimilate atmospheric CO2: C3, C4, and Crassulacean acid metabolism (CAM) [8][9] (Figure 1). CAM photosynthesis has evolved independently multiple times from C3 as a photosynthetic adaptation to cope with the decreasing atmospheric CO2 levels ~20 million years ago [10]. CAM plants are commonly found in harsh environments such as arid and semi-arid regions [11]. Other than those water-limited regions, CAM plants also inhabit the aquatic environment. With the release of the genome and transcriptome of an underwater CAM plant Isoetes taiwanensis [12], differences in the recruitment of phosphoenolpyruvate (PEP) carboxylase (PEPC) and core CAM pathway gene expression between aquatic and terrestrial plants demonstrate a different route of CAM evolution.
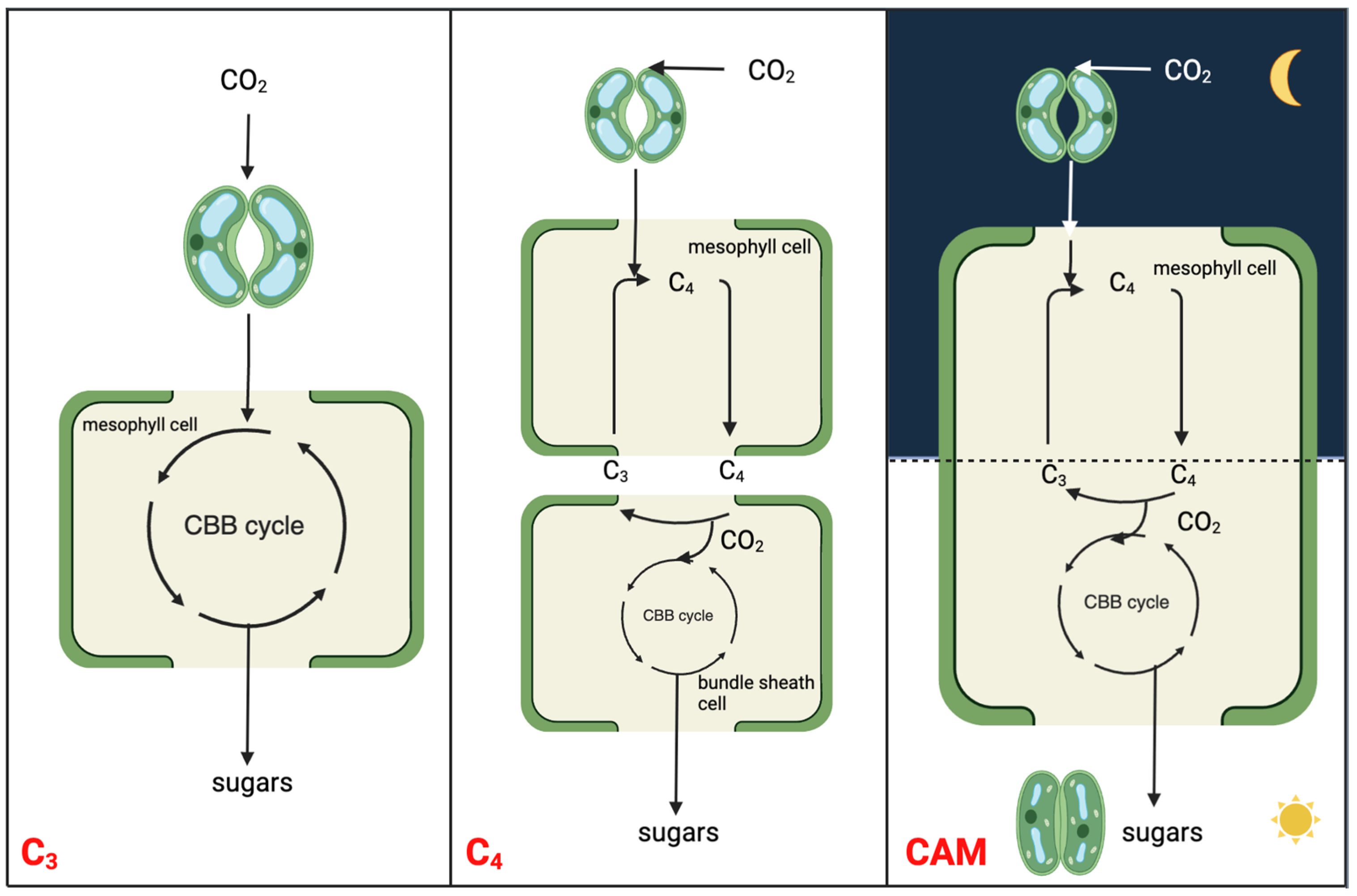
Figure 1. Simplified schematic to illustrate the molecular relationships and distinctions among C3, C4, and CAM photosynthesis mechanisms. The CBB cycle is the abbreviation of Calvin-Benson-Bassham cycle, which is also known as Calvin cycle.
CAM is a carbon concentrating mechanism, with the capability of assimilating CO2 initially at night using PEPC in the cytosol, leading to the formation of a four-carbon malate, which is then stored in the vacuole [13][14]. The three-carbon acceptor in this reaction is PEP, which is replenished by the glycolytic breakdown of carbohydrate storage in the form of starch or other sugars. Unlike spatial decoupling of carboxylation and decarboxylation in C4 photosynthesis, CAM photosynthesis separates these two processes in a temporal manner to shield ribulose-1,5-bisphosphate carboxylase-oxygenase (RuBisCO) from the oxygenase activity, minimizing photorespiration (Figure 1). CAM plants conduct gas exchange predominantly at night when the air temperature is low, thereby having a lower water loss by an order of magnitude than it would be during the day [15]. As such, CAM plants have water-use efficiency (WUE) several-fold higher than those of C3 and C4 plants under comparable conditions [16]. High WUE, together with enhanced heat and drought tolerance, drives the basic and applied research on CAM toward crop CAM engineering/bio-design. The typical diel cycle of CAM entails four phases: (I) nocturnal atmospheric CO2 fixation by PEPC and malic acid storage in the vacuole; (II) RuBisCO activation just after dawn when, for a brief period, CO2 is fixed by both PEPC and RuBisCO; (III) stored malate decarboxylated to CO2, which is fixed by RuBisCO; (IV) the end of the light period when stomata reopen driven by the depletion of malate pool [13][17].
CAM plants normally exhibit the following features: the diurnal fluctuation of malic acids (accumulation during the night period and dissipation during the day); reciprocal diurnal fluctuation of storage carbohydrates such as starch, polyglucans, or soluble hexoses; a high level of PEPC and an active decarboxylase; large storage vacuoles that are in the same cells with chloroplasts; water-limitation related traits, such as dense trichomes, leaf succulence, and waxy cuticles [1]; and nocturnal net CO2 uptake, which exhibits an inverse pattern of stomatal movement [18]. In the course of evolution, an intermediate CAM mechanism called facultative CAM arose. Plants conducting facultative CAM demonstrate the optional use of CAM under stress conditions, while remaining the use of C3 or C4 photosynthesis under normal conditions [19][20][21][22][23].
Given the characteristics of facultative CAM photosynthesis and climate change urgency, unraveling the molecular mechanisms underlying the C3 to CAM transition has attracted growing interest. However, there is a lack of review on facultative CAM and especially C3 to CAM transition. Herein resesarchers summarize recent advances in facultative C3 to CAM transition, discuss current problems and challenges, and highlight future research directions (Figure 2).
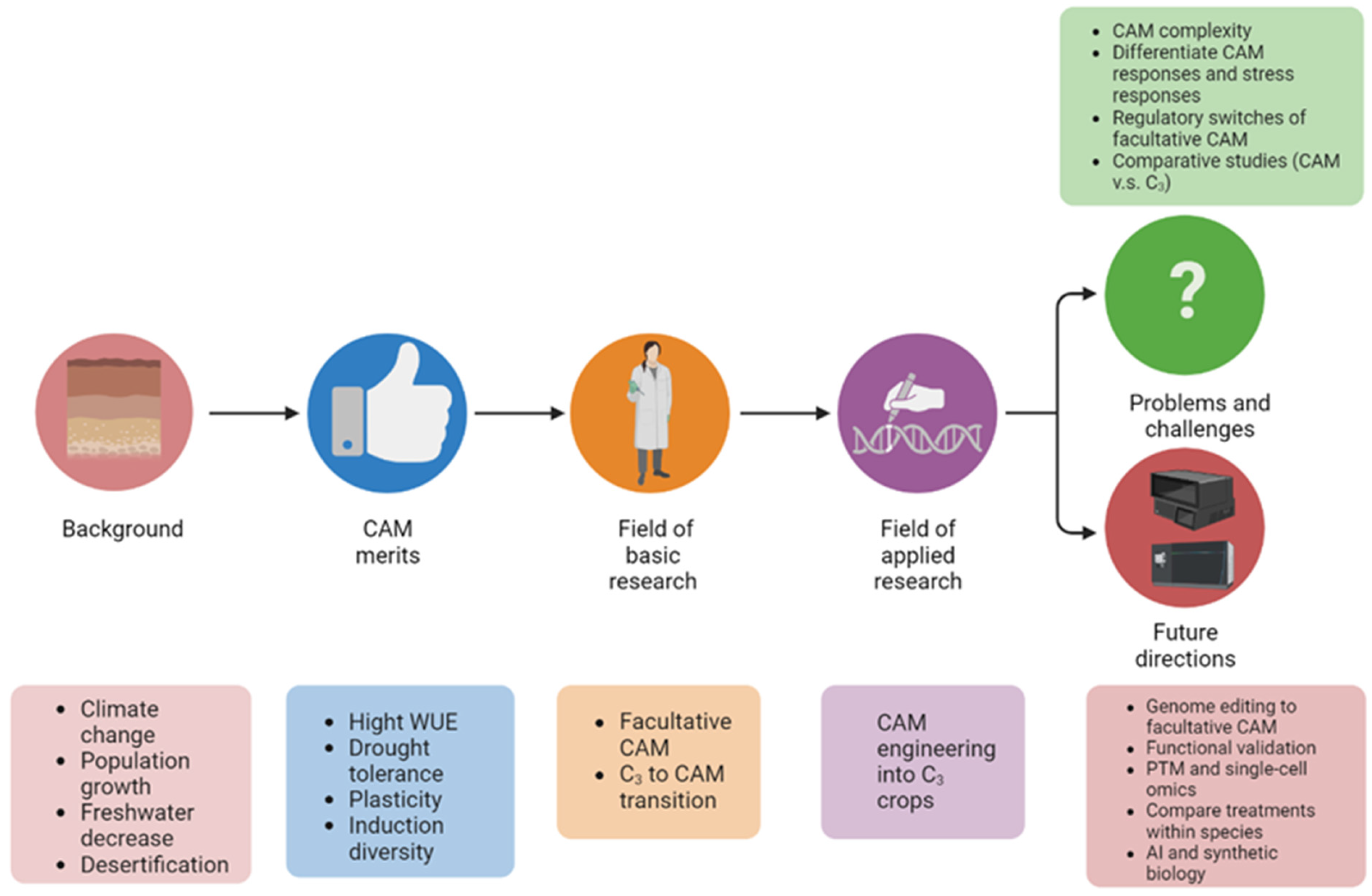
Figure 2. Graphical summary of the significance of studying C3 to CAM transition of facultative CAM plants. The abbreviations used: C3, C3 photosynthesis; CAM, Crassulacean acid metabolism; WUE, water-use efficiency; and PTM, post-translational modification.
2. The Plasticity of CAM Is Best Represented in Facultative CAM
A remarkable hallmark of CAM plants is their considerable plasticity in expressing the four phases of CAM, while keeping the C3 cycle fully functional [24]. Environmental factors such as light intensity, relative humidity, water availability [25], and developmental stages [16] affect the degree and duration of CAM expression [9]. CAM is highly plastic and can operate in different modes: (1) obligate/constitutive CAM or strong CAM, with high nocturnal acid accumulation (ΔH+) and CO2 fixation; (2) CAM-cycling, with daytime CO2 fixation like C3 and nocturnal fixation of CO2 from respiration; (3) CAM idling, with stomata closed all the time and CAM fixation of CO2 only from respiration; (4) facultative/inducible CAM, with C3 mode of CO2 fixation and zero ΔH+ in the non-stressed state, and small nocturnal CO2 fixation and ΔH+ during C3 to CAM transition in the stressed state [26]; (5) weak CAM, with similar CO2 uptake pattern as strong CAM but less nocturnal acid accumulation.
Among the above five different modes of CAM, a preeminent model for elucidating the molecular underpinnings of CAM is facultative CAM [27][28]. In facultative CAM species, CAM may be induced by a variety of stimuli such as drought [29][30], salinity [19][31], high photosynthetic photon flux [6][32], abscisic acid (ABA) [33], photoperiod [34] and hydrogen peroxide [35]. Clearly, CAM plasticity is best represented by facultative CAM plants, which employ the C3 photosynthesis under non-stress conditions to maximize growth, but are able to undergo a gradual C3 to CAM transition to reduce water loss and maintain photosynthetic integrity under water-limited conditions. It ultimately translates into high WUE, survival, and reproductive success [36]. Facultative CAM plants have been identified in a wide range of plant families, such as Bromeliaceae, Cactaceae, Aizoaceae, Montiaceae, Lamiaceae, Vitaceae, and Didiereaceae [37], indicating multiple independent evolutionary events (Figure 3). Whether these independent events generated similar or different genetic and epigenetic changes that enable facultative CAM deserves immediate investigation, e.g., by identifying and utilizing the evolutionary pairs of C3 and CAM species.
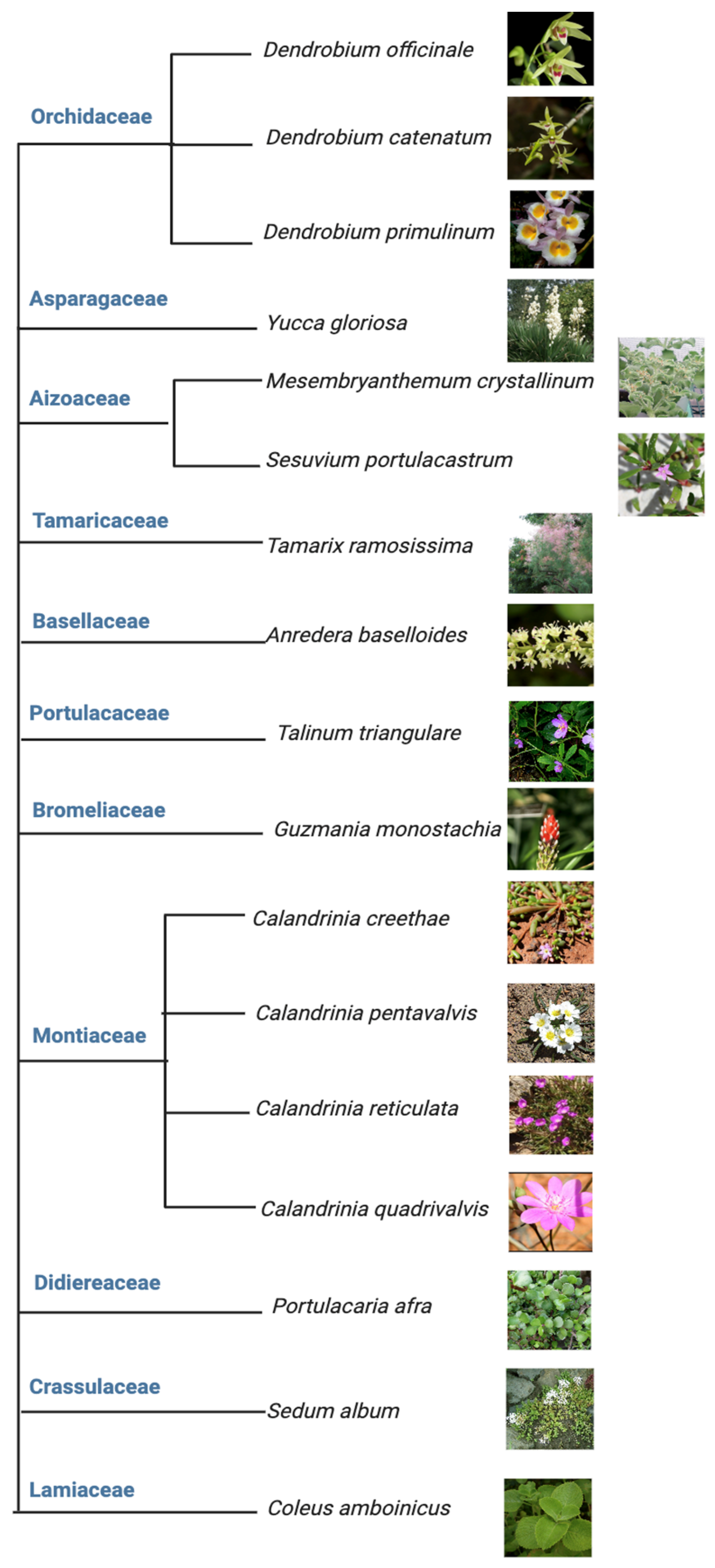
Figure 3. Tree view of all the facultative CAM plants investigated from 2017 to 2023.
3. Studies on C3 to CAM Transition Revealed Important Molecular Players
Over the past decades, different model species were used for diverse aspects of research pertinent to CAM. Here are six main areas of CAM research: (1) CAM ecophysiology to study and discover new CAM species in different ecological environments (e.g., [27][38]); (2) CAM origin and evolution (e.g., [39][40]); (3) genomic features and molecular mechanisms regulating CAM (e.g., [41][42]); (4) C3 to CAM transition (e.g., [19][20]); (5) CAM metabolic modeling (e.g., [43][44]); and (6) engineering CAM into C3 plants (e.g., [5][45]). All these areas of basic research aim for the ultimate goal of exploiting the potential of CAM in crop improvement under climate change [40]. Based on recent publications, Ananas comosus (Pineapple), Kalanchoë fedtschenkoi, and Mesembryanthemum crystallinum (Common ice plant, Table 1) are the three most extensively studied CAM models. Their genomes have been fully sequenced [42][46][47]. For studying the C3 to CAM transition, M. crystallinum has been the classic model, and Talinum triangulare is an emerging model [17]. T. triangulare is an herbaceous weed that shifts from C3 to CAM photosynthesis on day 11 of drought treatment. The large evenly green leaves, rapid growth, relatively short life cycle, self-cross, and full reversibility of CAM make it a model system to study facultative CAM [28].
Table 1. Research progress on CAM in M. crystallinum from 2017 to 2022.
| Research Focus | Key Findings | Reference |
|---|---|---|
| Identified C3-CAM transition period and temporal physiological changes |
|
[20] |
| Transcriptomics of guard cells during the C3-CAM transition |
|
[19] |
|
||
| Nocturnal carboxylation is coordinated with starch degradation by the products of these pathways, such as carbohydrates |
|
[48] |
| Functional CAM withdrawal in the de-salted plants |
|
[49] |
| Comparative proteomic changes in guard cells and mesophyll cells during the C3-CAM transition |
|
[50] |
| Phytohormones in the stomatal behavior during the C3-CAM transition |
|
[51] |
| Genome sequencing, transcriptomics, and comparative genomics of leaves |
|
[46] |
The early research of C3 to CAM transition mainly focused on several key CAM enzymes, such as PEPC [52], PEPC protein kinase (PPCK) and malic enzymes, as well as metabolite transport in the C3 and CAM state [53][54]. Early studies suggest that ABA signaling, Ca2+ signaling, and protein phosphorylation/de-phosphorylation may play important roles in the C3 to CAM transition, whereas no key players such as kinases/phosphatases were identified. Sequence analyses were also performed on PPC1 and PPC2, which are CAM-specific genes that encode PEPC [55][56]. With the emergence of microarray technologies, large-scale mRNA profiling was carried out [57]. Instead of studying the transition, Cushman group compared the gene expression of ice plants between the non-stressed C3 group and the induced-CAM group after 14 days of salt treatment. Gene expression of eight transporters was analyzed to study the inter-organellar metabolite transport between C3 and the CAM group of ice plants [58]. However, there is a lack of understanding of the temporal metabolic and molecular control of the C3 to CAM transition and a systems-level understanding was needed to reveal the regulatory changes underlying the transition.
With the advances in high-throughput omics technologies and computational biology, systems biology has become a prevalent approach for discovery (hypothesis generation) and functional studies (hypothesis testing). Beyond traditional physiological and biochemical methods, multi-omics (genomics, transcriptomics, proteomics and metabolomics) has generated a systems-level understanding of temporal molecular and metabolic controls underpinning CAM [59]. In T. triangulare leaves, targeted metabolite profiling and RNA sequencing were performed to reveal the rewiring of carbohydrate metabolism and candidate transcription factors (TFs) in the drought-induced CAM transition process [60]. Three years later, the same group identified seven candidate regulators of ABA-induced CAM including HEAT SHOCK TF A2, NUCLEAR FACTOR Y, SUBUNITS A9, and JmjC DOMAIN-CONTAINING PROTEIN 27 [61]. In addition to the traditional drought and salt induction of CAM, hydrogen peroxide was shown to be able to induce CAM in M. crystallinum [35]. In the leaves of another facultative bromeliad Guzmania monostachia, increases in the expression of CAM-related genes (PEPC1, PPCK, NAD-malate dehydrogenase, aluminum-activated malate transporter 9 (ALMT9), PEP carboxykinase (PEPCK)) and UREASE transcripts were shown under drought. And the role of integrating N and C metabolism of urea was suggested [62]. The CAM gene expression, antioxidant activities, and chlorophyll fluorescence were compared between a C3-CAM facultative species (Sedum album) and a C4-CAM facultative species (Portulaca oleracea) [63]. The level of nitric oxide (NO) was found to be correlated with the CAM expression during CAM induction only in S. album but not P. oleracea. This suggests the different roles of NO in C3 and C4 species during CAM induction. All the aforementioned studies did not identify a critical transition period, which is key to capturing the molecular switches for CAM. Three years ago, the transition period of M. crystallinum was first defined during salt-induced C3 to CAM shift, and further validated in independent studies through RNA-seq and physiological analyses [19][20][51]. Interestingly, three phytohormones, jasmonic acid (JA), cytokinin, and ABA were reported to play important roles in the inversed pattern of stomata opening/closing during the transition of M. crystallinum [51]. With the release of the ice plant genome [46], more studies can explore the genes and metabolites pertinent to the C3 to CAM transition.
This entry is adapted from the peer-reviewed paper 10.3390/ijms241713072
References
- Leisner, C.P. Review: Climate change impacts on food security- focus on perennial cropping systems and nutritional value. Plant Sci. 2020, 293, 110412.
- Reidmiller, D.R.; Avery, C.W.; Easterling, D.R.; Kunkel, K.E.; Lewis, K.L.M.; Maycock, T.K.; Stewart, B.C. (Eds.) USGCRP Impacts, Risks, and Adaptation in the United States: Fourth National Climate Assessment; U.S. Global Change Research Program: Washington, DC, USA, 2018; Volume II, p. 1515.
- Yao, J.; Liu, H.; Huang, J.; Gao, Z.; Wang, G.; Li, D.; Yu, H.; Chen, X. Accelerated dryland expansion regulates future variability in dryland gross primary production. Nat. Commun. 2020, 11, 1665.
- Eshel, G.; Araus, V.; Undurraga, S.; Soto, D.C.; Moraga, C.; Montecinos, A.; Moyano, T.; Maldonado, J.; Díaz, F.P.; Varala, K.; et al. Plant ecological genomics at the limits of life in the Atacama Desert. Proc. Natl. Acad. Sci. USA 2021, 118, e2101177118.
- Lim, S.D.; Mayer, J.A.; Yim, W.C.; Cushman, J.C. Plant tissue succulence engineering improves water-use efficiency, water-deficit stress attenuation and salinity tolerance in Arabidopsis. Plant J. 2020, 103, 1049–1072.
- Hu, R.; Zhang, J.; Jawdy, S.; Sreedasyam, A.; Lipzen, A.; Wang, M.; Ng, V.; Daum, C.; Keymanesh, K.; Liu, D.; et al. Comparative genomics analysis of drought response between obligate CAM and C3 photosynthesis plants. J. Plant Physiol. 2022, 277, 153791.
- Blankenship, R.E. Molecular Mechanisms of Photosynthesis; Wiley-Blackwell: Hoboken, NJ, USA, 2008; pp. 1–10.
- Crayn, D.M.; Winter, K.; Smith, J.A.C. Multiple origins of crassulacean acid metabolism and the epiphytic habit in the Neotropical family Bromeliaceae. Proc. Natl. Acad. Sci. USA 2004, 101, 3703–3708.
- Ceusters, J.; Van De Poel, B. Ethylene Exerts Species-Specific and Age-Dependent Control of Photosynthesis. Plant Physiol. 2018, 176, 2601–2612.
- Heyduk, K. Evolution of Crassulacean acid metabolism in response to the environment: Past, present, and future. Plant Physiol. 2022, 190, 19–30.
- Amin, A.B.; Rathnayake, K.N.; Yim, W.C.; Garcia, T.M.; Wone, B.; Cushman, J.C.; Wone, B.W.M. Crassulacean Acid Metabolism Abiotic Stress-Responsive Transcription Factors: A Potential Genetic Engineering Approach for Improving Crop Tolerance to Abiotic Stress. Front. Plant Sci. 2019, 10, 129.
- Wickell, D.; Kuo, L.-Y.; Yang, H.-P.; Ashok, A.D.; Irisarri, I.; Dadras, A.; de Vries, S.; de Vries, J.; Huang, Y.-M.; Li, Z.; et al. Underwater CAM photosynthesis elucidated by Isoetes genome. Nat. Commun. 2021, 12, 6348.
- Osmond, C.B. Crassulacean Acid Metabolism: A Curiosity in Context. Annu. Rev. Plant Physiol. 1978, 29, 379–414.
- Borland, A.M.; Guo, H.-B.; Yang, X.; Cushman, J.C. Orchestration of carbohydrate processing for crassulacean acid metabolism. Curr. Opin. Plant Biol. 2016, 31, 118–124.
- Bowyer, J.; Leegood, R. Photosynthesis. In Plant Biochemistry; Dey, P.M., Harborne, J.B., Eds.; Academic Press: London, UK, 1997; pp. 1–4, 49–110.
- Drennan, P.M.; Nobel, P.S. Responses of CAM species to increasing atmospheric CO2 concentrations. Plant Cell Environ. 2000, 23, 767–781.
- Schiller, K.; Bräutigam, A. Engineering of Crassulacean Acid Metabolism. Annu. Rev. Plant Biol. 2021, 72, 77–103.
- Kluge, M.; Ting, I.P. Crassulacean Acid Metabolism; Springer: Berlin/Heidelberg, Germany, 1978; pp. 108–152.
- Kong, W.; Yoo, M.J.; Zhu, D.; Noble, J.D.; Kelley, T.M.; Li, J.; Kirst, M.; Assmann, S.M.; Chen, S. Molecular changes in Mesembryanthemum crystallinum guard cells underlying the C3 to CAM transition. Plant Mol. Biol. 2020, 103, 653–667.
- Guan, Q.; Tan, B.; Kelley, T.M.; Tian, J.; Chen, S. Physiological changes in Mesembryanthemum crystallinum during the C3 to CAM transition induced by salt stress. Front. Plant Sci. 2020, 11, 283.
- Ferrari, R.C.; Kawabata, A.B.; Ferreira, S.S.; Hartwell, J.; Freschi, L. A matter of time: Regulatory events behind the synchronization of C4 and crassulacean acid metabolism in Portulaca oleracea. J. Exp. Bot. 2022, 73, 4867–4885.
- Gilman, I.S.; Moreno-Villena, J.J.; Lewis, Z.R.; Goolsby, E.W.; Edwards, E.J. Gene co-expression reveals the modularity and integration of C4 and CAM in Portulaca. Plant Physiol. 2022, 189, 735–753.
- Ferrari, R.C.; Bittencourt, P.P.; Rodrigues, M.A.; Moreno-Villena, J.J.; Alves, F.R.R.; Gastaldi, V.D.; Boxall, S.F.; Dever, L.V.; Demarco, D.; Andrade, S.C.S.; et al. C4 and crassulacean acid metabolism within a single leaf: Deciphering key components behind a rare photosynthetic adaptation. New Phytol. 2020, 225, 1699–1714.
- Gilman, I.S.; Edwards, E.J. Crassulacean acid metabolism. Curr. Biol. 2020, 30, R57–R62.
- Cushman, J.C. Crassulacean acid metabolism. A plastic photosynthetic adaptation to arid environments. Plant Physiol. 2001, 127, 1439–1448.
- Herrera, A.; Martin, C.E.; Tezara, W.; Ballestrini, C.; Medina, E. Induction by drought of Crassulacean acid metabolism in the terrestrial bromeliad, Puya floccose. Photosynthetica 2010, 48, 383–388.
- Winter, K. Ecophysiology of constitutive and facultative CAM photosynthesis. J. Exp. Bot. 2019, 70, 6495–6508.
- Winter, K.; Holtum, J.A.M. Facultative Crassulacean acid metabolism (CAM) plants: Powerful tools for unravelling the functional elements of CAM photosynthesis. J. Exp. Bot. 2014, 65, 3425–3441.
- Borland, A.M.; Griffiths, H. The regulation of CAM and respiratory recycling by water supply and light regime in the C3 -CAM intermediate Sedum telephium. Funct. Ecol. 1990, 4, 33.
- Olivares, E.; Urich, R.; Montes, G.; Coronel, I.; Herrera, A. Occurrence of Crassulacean acid metabolism in Cissus trifoliata L. (Vitaceae). Oecologia 1984, 61, 358–362.
- Winter, K.; von Willert, D.J. NaCl-induzierter crassulaceensäurestoffwechsel bei Mesembryanthemum crystallinum. Z. Pflanzenphysiol. 1972, 67, 166–170.
- Maxwell, K. Resistance is useful: Diurnal patterns of photosynthesis in C3 and Crassulacean acid metabolism epiphytic bromeliads. Funct. Plant Biol. 2002, 29, 679–687.
- Taybi, T.; Cushman, J.C. Abscisic acid signaling and protein synthesis requirements for phosphoenolpyruvate carboxylase transcript induction in the common ice plant. J. Plant Physiol. 2002, 159, 1235–1243.
- Brulfert, J.; Kluge, M.; Güçlü, S.; Queiroz, O. Interaction of photoperiod and drought as CAM inducing factors in Kalanchoë blossfeldiana Poelln., cv. Tom Thumb. J. Plant Physiol. 1988, 133, 222–227.
- Surówka, E.; Dziurka, M.; Kocurek, M.; Goraj, S.; Rapacz, M.; Miszalski, Z. Effects of exogenously applied hydrogen peroxide on antioxidant and osmoprotectant profiles and the C3-CAM shift in the halophyte Mesembryanthemum crystallinum L. J. Plant Physiol. 2016, 200, 102–110.
- Winter, K.; Ziegler, H. Induction of Crassulacean acid metabolism in Mesembryanthemum crystallinum increases reproductive success under conditions of drought and salinity stress. Oecologia 1992, 92, 475–479.
- Nosek, M.; Gawrońska, K.; Rozpądek, P.; Sujkowska-rybkowska, M.; Miszalski, Z.; Kornaś, A. At the edges of photosynthetic metabolic plasticity—On the rapidity and extent of changes accompanying salinity stress-induced cam photosynthesis withdrawal. Int. J. Mol. Sci. 2021, 22, 8426.
- Winter, K.; Garcia, M.; Virgo, A.; Smith, J.A.C. Low-level CAM photosynthesis in a succulent-leaved member of the Urticaceae, Pilea peperomioides. Funct. Plant Biol. 2021, 48, 683–690.
- Bräutigam, A.; Schlüter, U.; Eisenhut, M.; Gowik, U. On the evolutionary origin of CAM photosynthesis. Plant Physiol. 2017, 174, 473–477.
- Yang, X.; Cushman, J.C.; Borland, A.M.; Edwards, E.J.; Wullschleger, S.D.; Tuskan, G.A.; Owen, N.A.; Griffiths, H.; Smith, J.A.C.; De Paoli, H.C.; et al. A roadmap for research on crassulacean acid metabolism (CAM) to enhance sustainable food and bioenergy production in a hotter, drier world. New Phytol. 2015, 207, 491–504.
- Boxall, S.F.; Kadu, N.; Dever, L.V.; Knerová, J.; Waller, J.L.; Gould, P.J.D.; Hartwell, J. Kalanchoë PPC1 is essential for Crassulacean acid metabolism and the regulation of core circadian clock and guard cell signaling genes. Plant Cell 2020, 32, 1136–1160.
- Yang, X.; Hu, R.; Yin, H.; Jenkins, J.; Shu, S.; Tang, H.; Liu, D.; Weighill, D.A.; Yim, W.C.; Ha, J.; et al. The Kalanchoë genome provides insights into convergent evolution and building blocks of crassulacean acid metabolism. Nat. Commun. 2017, 8, 1899.
- Shameer, S.; Baghalian, K.; Cheung, C.Y.M.; Ratcliffe, R.G.; Sweetlove, L.J. Computational analysis of the productivity potential of CAM. Nat. Plants 2018, 4, 165–171.
- Tay, I.Y.Y.; Odang, K.B.; Cheung, C.Y.M. Metabolic modeling of the C3-CAM continuum revealed the establishment of a starch/sugar-malate cycle in CAM evolution. Front. Plant Sci. 2021, 11, 573197.
- Lim, S.D.; Lee, S.; Choi, W.G.; Yim, W.C.; Cushman, J.C. Laying the foundation for Crassulacean acid metabolism (CAM) biodesign: Expression of the C4 metabolism cycle genes of CAM in Arabidopsis. Front. Plant Sci. 2019, 10, 101.
- Shen, S.; Li, N.; Wang, Y.; Zhou, R.; Sun, P.; Lin, H.; Chen, W.; Yu, T.; Liu, Z.; Wang, Z.; et al. High-quality ice plant reference genome analysis provides insights into genome evolution and allows exploration of genes involved in the transition from C3 to CAM pathways. Plant Biotechnol. J. 2022, 20, 2107–2122.
- Ming, R.; VanBuren, R.; Wai, C.M.; Tang, H.; Schatz, M.C.; Bowers, J.E.; Lyons, E.; Wang, M.-L.; Chen, J.; Biggers, E.; et al. The pineapple genome and the evolution of CAM photosynthesis. Nat. Genet. 2015, 47, 1435–1442.
- Taybi, T.; Cushman, J.C.; Borland, A.M. Leaf carbohydrates influence transcriptional and post-transcriptional regulation of nocturnal carboxylation and starch degradation in the facultative CAM plant, Mesembryanthemum crystallinum. J. Plant Physiol. 2017, 218, 144–154.
- Nosek, M.; Gawrońska, K.; Rozpądek, P.; Szechyńska-Hebda, M.; Kornaś, A.; Miszalski, Z. Withdrawal from functional Crassulacean acid metabolism (CAM) is accompanied by changes in both gene expression and activity of antioxidative enzymes. J. Plant Physiol. 2018, 229, 151–157.
- Guan, Q.; Kong, W.; Zhu, D.; Zhu, W.; Dufresne, C.; Tian, J.; Chen, S. Comparative proteomics of Mesembryanthemum crystallinum guard cells and mesophyll cells in transition from C3 to CAM. J. Proteom. 2021, 231, 104019.
- Wakamatsu, A.; Mori, I.C.; Matsuura, T.; Taniwaki, Y.; Ishii, R.; Yoshida, R. Possible roles for phytohormones in controlling the stomatal behavior of Mesembryanthemum crystallinum during the salt-induced transition from C3 to Crassulacean acid metabolism. J. Plant Physiol. 2021, 262, 153448.
- Winter, K. Properties of phosphoenolpyruvate carboxylase in rapidly prepared, desalted leaf extracts of the Crassulacean acid metabolism plant Mesembryanthemum crystallinum L. Planta 1982, 154, 298–308.
- Häusler, R.E.; Baur, B.; Scharte, J.; Teichmann, T.; Eicks, M.; Fischer, K.L.; Flügge, U.I.; Schubert, S.; Weber, A.; Fischer, K. Plastidic metabolite transporters and their physiological functions in the inducible crassulacean acid metabolism plant Mesembryanthemum crystallinum. Plant J. 2000, 24, 285–296.
- Neuhaus, H.E.; Holtum, J.A.; Latzko, E. Transport of phosphoenolpyruvate by chloroplasts from Mesembryanthemum crystallinum L. exhibiting Crassulacean acid metabolism. Plant Physiol. 1988, 87, 64–68.
- Cushman, J.C.; Meyer, G.; Michalowski, C.B.; Schmitt, J.M.; Bohnert, H.J. Salt stress leads to differential expression of two isogenes of phosphoenolpyruvate carboxylase during Crassulacean acid metabolism induction in the common ice plant. Plant Cell 1989, 1, 715–725.
- Vaasen, A.; Begerow, D.; Hampp, R. Phosphoenolpyruvate carboxylase genes in C3, Crassulacean acid metabolism (CAM) and C3/CAM intermediate species of the genus Clusia: Rapid reversible C3/CAM switches are based on the C3 housekeeping gene. Plant Cell Environ. 2006, 29, 2113–2123.
- Cushman, J.C.; Tillett, R.L.; Wood, J.A.; Branco, J.M.; Schlauch, K.A. Large-scale mRNA expression profiling in the common ice plant, Mesembryanthemum crystallinum, performing C3 photosynthesis and Crassulacean acid metabolism (CAM). JXB 2008, 59, 1875–1894.
- Kore-Eda, S.; Noake, C.; Ohishi, M.; Ohnishi, J.I.; Cushman, J.C. Transcriptional profiles of organellar metabolite transporters during induction of Crassulacean acid metabolism in Mesembryanthemum crystallinum. Funct. Plant Biol. 2005, 32, 451–466.
- Abraham, P.E.; Yin, H.; Borland, A.M.; Weighill, D.; Lim, S.D.; De Paoli, H.C.; Engle, N.; Jones, P.C.; Agh, R.; Weston, D.J.; et al. Transcript, protein and metabolite temporal dynamics in the CAM plant Agave. Nat. Plants 2016, 2, 16178.
- Brilhaus, D.; Bräutigam, A.; Mettler-Altmann, T.; Winter, K.; Weber, A.P. Reversible burst of transcriptional changes during induction of Crassulacean acid metabolism in Talinum triangulare. Plant Physiol. 2016, 170, 102–122.
- Maleckova, E.; Brilhaus, D.; Wrobel, T.J.; Weber, A.P.M. Transcript and metabolite changes during the early phase of abscisic acid-mediated induction of Crassulacean acid metabolism in Talinum triangulare. JXB 2019, 70, 6581–6596.
- Gonçalves, A.Z.; Mercier, H. Transcriptomic and biochemical analysis reveal integrative pathways between carbon and nitrogen metabolism in Guzmania monostachia (Bromeliaceae) under drought. Front. Plant Sci. 2021, 12, 715289.
- Habibi, G. Comparison of CAM expression, photochemistry and antioxidant responses in Sedum album and Portulaca oleracea under combined stress. Physiol. Plant 2020, 170, 550–568.
This entry is offline, you can click here to edit this entry!
