Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Biochemistry & Molecular Biology
Human whole saliva is a hypotonic fluid lining the oral cavity and is composed of water (99%) and a complex mixture of organic and inorganic compounds resulting from salivary gland secretion, oral flora, the oropharynx, the upper airway, gastrointestinal reflux, gingival crevicular fluid, food deposits, and mucosal surface secretion containing blood-derived components. The most-common PTM detectable in human saliva is the proteolytic cleavage of proteins.
- salivary proteins
- post-translational modifications
1. Introduction
Human whole saliva is a hypotonic fluid lining the oral cavity and is composed of water (99%) and a complex mixture of organic and inorganic compounds resulting from salivary gland secretion, oral flora, the oropharynx, the upper airway, gastrointestinal reflux, gingival crevicular fluid, food deposits, and mucosal surface secretion containing blood-derived components [1]. Saliva is essential for the accomplishment of multiple physiological functions, encompassing lubrication, buffering, the maintenance of tooth integrity, chewing, the initial digestion of some foods, swallowing, tissue hydration and lubrication, speech, and wound healing, and it exhibits antibacterial and antifungal activity [2]. The dramatic sequelae observed in patients suffering from Sjögren syndrome clearly demonstrate the relevance of saliva and its components, particularly salivary proteins, in the protection of the mouth. Recent proteomic inventories report more than 3000 proteoforms in human saliva [3]. Before, during, and after secretion, most salivary proteins undergo numerous post-translational modifications (PTMs), of which the roles have not yet been clearly elucidated. Over the last twenty-five years, researchers' group has been able to characterize many PTMs of the salivary proteome, mainly by applying top-down proteomic platforms to saliva from healthy subjects of various ages and from patients affected by various pathologies.
2. Enzymatic Cleavages (Cryptides)
The most-common PTM detectable in human saliva is the proteolytic cleavage of proteins. Indeed, most salivary proteins are submitted to the action of endogenous and exogenous proteinases (the last mainly from oral flora), which leads to the formation of a myriad of fragments. Many of these fragments could be considered important members of the cryptide family, defined as bioactive peptides encrypted inside a bigger functional polypeptide and released by a proteolytic event, with distinct or related function, but not superimposable, to that of the parental polypeptides [4].
2.1. Pre-Secretory Cleavages
The secretory pathway for many proteins includes transit in the Golgi apparatus and storage in secretory granules preceding their release from the cell into the duct system and secretion into the mouth [5]. Most of the pre-secretory cleavages of proteins occur during the transport towards the granules of the trans-Golgi-network [6], which represents the major secretory-pathway sorting station. Glandular secretions, protein extracts from secretory granules isolated from the major salivary glands, and whole saliva were investigated by top-down proteomics to characterize and distinguish between events occurring prior to the storage in the secretory granules, those taking place between granule release and secretion into the mouth during the passage through the secretory duct, and those occurring into the mouth [6]. The workflow applied in that study is represented in Figure 1. Proteases involved in the most relevant cleavages belong to the convertase and exopeptidase families [6,7,8,9].
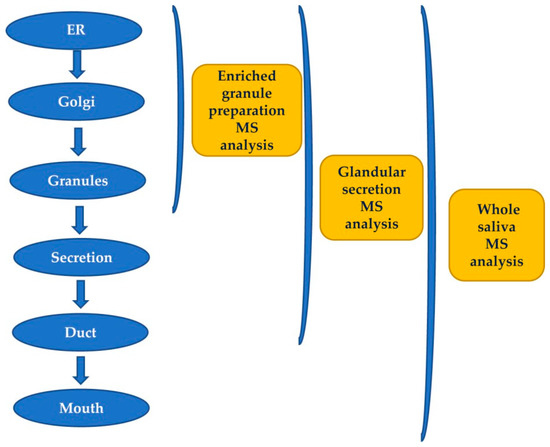
Figure 1. Workflow applied for studying the origin of post-translational modifications (PTMs) of salivary peptides and proteins secreted by the major salivary glands by a top-down proteomic approach.
2.1.1. Proline-Rich Proteins (PRPs)
Human PRPs constitute a very polymorphic family of proteins, missing amino acids in their sequence aromatic and characterized by a high content of proline (as well as glutamine and glycine). All PRPs are encoded by chromosome 12p13.2 and are divided into three classes based on their ionic properties, i.e., acidic proline-rich-proteins (aPRPs), encoded by PRH1 and PRH2 loci; basic proline-rich proteins (bPRPs), encoded by the PRB1, PRB2, and PRB4 loci located (considering the direction of translation) immediately before the aPRPs loci; and the glycosylated (basic) proline-rich proteins (gPRPs), encoded by the PRB3 locus, which is the fourth locus of the bPRPs cluster. It is relevant to underline that except for the bPRPs named P-D, the other three proteoforms of the PRB-4 locus, i.e., the IB-8a (Con 1+) of PRB-2 and all the products of the PRB-3 locus, are submitted to glycosylation, which will be described in more detail, if available, in the section devoted to this PTM.
Acidic Proline-Rich-Proteins (aPRPs)
aPRPs exist as five principal isoforms called PRP-1, PRP-2 (coded by PRH-2 locus) and Pif-s (parotid isoelectric-focusing isoform-slow), Pa (parotid acidic isoform), and Db-s (double band isoform-slow) (coded by PRH-1 locus). The isoforms PRP-1, PRP-2, Pif-s, and Pa are all 150 residues long. Taking as reference PRP-2, the Pif-s and Pa isoforms have an asparagine (Asn4) instead of Asp4, and PRP-1 has Asn50 instead of the Asp50 present in the other isoforms (Figure 2). The Db-s isoform is 171 a.a. residues long due to the insertion of 21 residues repeated after position 81 and, as Pa, it has a Leu27 instead of Ileu27. The PRP-1, PRP-2, and PIF-s isoforms are partially cleaved at the Arg106 by a convertase recognizing the …R103XXR106… consensus sequence, generating the PRP-3, PRP-4, and Pif-f (f stands for fast electrophoretic isoform) truncated isoforms and a terminal fragment of 44 amino-acid residues called the P-C (or IB-8b) peptide. The convertase consensus sequence of Db-s is shifted by the insertion, and it is cleaved at Arg127, generating the Db-f isoform and always the P-C peptide. The consensus sequence of the convertase responsible for the cleavage is preserved because the Pa isoform, having a Cys103 residue instead of Arg103, is not cleaved. Indeed, a truncated Pa isoform was never detected. Rather, it generates in the mouth a Pa-2-mer isoform due to the formation of a disulfide bridge between two Cys103 residues (Figure 2), which is the main Pa component detectable in whole saliva [9]. The cleavage of PRP-1, PRP-2, Pif-s, and Db-s is not complete, and in whole saliva, the entire and truncated isoforms are both detectable, approximatively in a proportion of 70/30%, respectively. All the entire isoforms are the PRP-1 type, while all the truncated are the PRP-3 type (obviously non-including the Pa proteoform). All these characteristics of aPRPs are summarized in Figure 2.
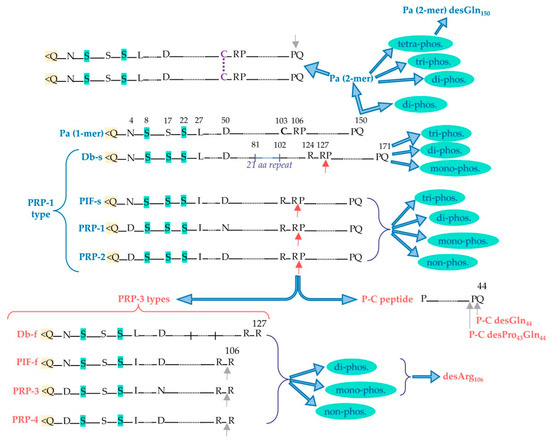
Figure 2. Principal acidic proline-rich protein (aPRP) proteoforms: in blue are the entire proteoforms and in red are the truncated ones. Red arrows indicate the cleavage site recognized by convertase activity, from which PRP-3-type truncated forms and the P-C peptide are generated from the entire PRP-1-type forms. Grey arrows indicate the possible loss of C-terminal residue by carboxypeptidase activity. Pyroglutamination N-terminal is evidenced in yellow, while the Ser residues that can undergo phosphorylation are evidenced in light green. The different phosphorylated proteoforms obtainable from the entire and truncated aPRPs are shown, as well as the disulfide dimeric form of Pa.
Basic-Proline-Rich-Proteins (bPRPs)
bPRPs are encoded by the polymorphic PRB1, PRB2, and PRB4 loci, existing as three or four possible alleles: (S(mall), M(edium), and L(arge), or for the eventual fourth allele, V(ery)L(arge)). Unlike aPRPs, they are completely cleaved by convertases during granule storage [6]. The proteoforms generated from the pre-pro-proteins are listed below:
-
Products of Locus PRB-1: II-2, P-E (or IB-9), P-Ko, IB-6, Ps-1, Ps-2.
-
Products of Locus PRB-2: IB-1, P-J, P-H (or IB-4), P-F (orIB-8c), IB-8a(Con 1+), IB-8a(Con 1−).
-
Products of Locus PRB-4: PGA, II-1, Cd-IIg, P-D (or IB-5).
The nomenclature reported for bPRPs is difficult and confusing since it derives from two different criteria. Kauffmann and colleagues [10,11,12,13] have purified eleven bPRPs and determined the sequence of ten of them. They named these proteoforms using the names of the different fractions obtained from an articulate multidimensional preparative chromatographic separation of whole saliva. In the same years, Saitoh and colleagues [14,15,16] and Isemura and colleagues [17,18] identified nine bPRPs, naming them from P-A to P-I, and sequenced seven of them. As reported in the list, the two nomenclatures have several overlaps. P-A and P-I are artifacts deriving from the proteolytic cleavage occurring during the purification of salivary proteins. Although included in the class of basic PRPs, P-B and P-C peptides are not codified by PRB1-PRB4 genes. Indeed, the P-B peptide is encoded by the PROL3 gene (PBI) clustered on chromosome 4q13.317. It is related to statherin and, therefore, it cannot be considered to pertain to the bPRP family. As described in the previous section on aPRPs, the P-C peptide derives from the cleavage of four isoforms of aPRPs, namely PRP-1, PRP-2, Pif-s, and Db-s. In a top-down MS proteomic study, researchers were able to characterize a new bPRP deriving from the PRB-2 locus, which was named P-J [19,20].
The pre-secretory cleavage of bPRP pro-proteins usually occurs at the level of the consensus sequence KSRSXR↓, where X may be Pro, Ser, or Ala [6,21,22]. An extensive characterization of the different proteoforms and fragments of bPRPs has been recently published [23], where the interested reader can find the list of the bPRPs components detected until 2018. In this inventory, a new classification of bPRPs was proposed based on the similarities of their sequence, dividing them into three groups, reported in Figure 3a–c. The first group, which researchers named Group 1, includes P-E, P-Ko, IB-6, Ps-1, Ps-2, P-H, P-F, P-J, and P-D (Figure 3a). The sequence of all these bPRPs starts with the same SPPGKPQGPP motif, followed by sequences somewhat similar but showing small variations among the different components. The central part of the sequences shows similar repeats. Because P-E, IB-6, Ps-1, and Ps-2 sequences originate from DNA-length polymorphisms in exon 3 of the PRB1 locus, they exhibit high similarity. The bPRP with a Mav of 10,433.5 Da, detected in whole saliva and in parotid secretory granules and named P-Ko, is encoded by cP4, a differentially spliced transcript of the PRB1-L allele.
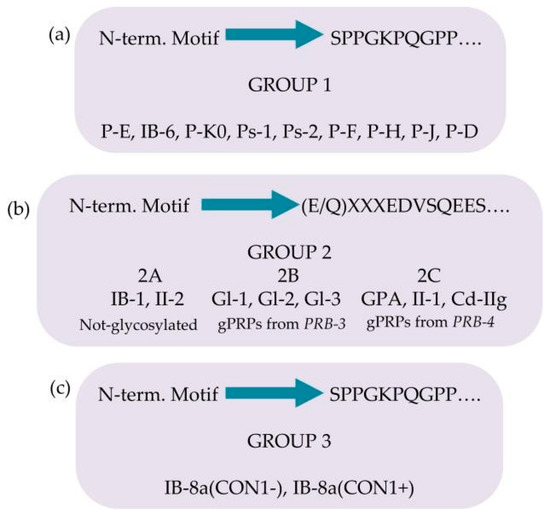
Figure 3. Classification of the basic proline-rich proteins (bPRPs) based on the similarity of the N-terminal motif and on the presence of glycosylations. Group 1 in panel (a), group 2 in panel (b), group 3 in panel (c).
Group 2 includes IB-1, II-2, and the glycosylated bPRPs codified by PRB3 and PRB4 genes, namely, Gl-1, Gl-2, Gl-3, GPA, II-1, and Cd-IIg (Figure 3b). Their sequences start with a similar motif (E/Q)XXXEDVSQEES, where XXX is LNE in IB-1, II-2, Gl-1, Gl-2, and Gl-3 and SSS in GPA, II-1, and Cd-IIg). The central part of the sequences comprises similar repeats, with differences from the repeats of the members belonging to Group 1. Based on structural differences and similarities, members of Group 2 can be divided into three subgroups: Group 2A, including IB-1 and II-2, without glycosylation sequons; Group 2B, including the Gl proteins codified by the alleles of PRB3 locus; and Group 2C, including the glycosylated proteins codified by the alleles of PRB4 locus. The Small Group (3) is a hybrid group, which includes the two proteoforms of IB-8a, Con1−, and Con1+ (Figure 3c). The initial sequence of these two proteins resembles that of Group 1, while the terminal sequence is like the repeat responsible for the glycosylation of the bPRPs of Groups 2B and 2C [23].
2.1.2. Further Pre-Secretory Cleavages of PRPs
All the PRP families described above are substrates of a carboxypeptidase, which removes the C-terminal residue and commonly, obviously, is an arginine. However, the enzyme is not specific. It removes other residues, and sometimes a second C-terminal loss is detectable [22]. The C-terminal pre-secretory cleavages occurring on PRPs are reported in Table 1.
Table 1. Some terminal losses detected in salivary peptides/proteins.
| Protein and Peptides | C-Term. Cleavage |
|---|---|
| aPRP Pa-dimer | …QSP↓Q |
| PRP3/PRP4/PIF-f | …RPP↓R |
| P-C (or IB-8b) | …QS↓P↓Q |
| bPRP II-2 | …RSP↓R |
| bPRP IB-1 | …RSP↓R |
| bPRP PF (or IB-8c) precursor | …RSA↓R |
| bPRP PE (or IB-9) | …RSP↓R |
2.1.3. Role of PRPs
Salivary proline-rich proteins are highly conserved in mammalian saliva [24]. Nonetheless, significant structural differences are present in the mammal families, suggesting that they play a relevant role in oral protection, in the modulation of the activity of oral ions, in the colonization of oral microbiota and the gastrointestinal tract, and in the feeding habit [24]. However, while aPRPs and the gPRP, called Gl 3M, are detectable in the saliva of preterm newborns, the other bPRPs are not detectable in human saliva until puberty [20,25,26], suggesting either a role in the perception of the taste of foods or a function in secondary sexual maturation. As we are aware, this information is not available for other mammals. In humans, bPRPs are secreted only by parotid glands, and this regioselectivity is also puzzling. Some bPRPs exhibit the ability to bind harmful tannins [27], others can modulate the oral flora [28,29], and some others are involved in bitter taste perception [30]. Some bPRP fragments are involved in enamel pellicle formation [31] and others act as antagonists of progesterone-induced cytosolic Ca2+ mobilization [32].
The intrinsic propensity of some fragments to adopt a polyproline-II helix arrangement joined to PXXP motifs was suggestive of the interaction with the SH3 domain family [33]. Interestingly, interactions were highlighted with Fyn, Hck, and c-Src SH3 domains [34], which are included in the Src kinases family, suggesting that some basic bPRPs can be involved in the signal transduction pathways modulated by these kinases. Only a small amount of data on correlations between genes of bPRPs and diseases linked to their allelic variants has been reported so far. In fact, for some of the alleles (PRB1VL, PRB2S, M, VL, and PRB3VL) the genetic sequence is not reported. Moreover, for the small and large alleles of PRB1, the genetic sequence is incomplete [35] because the reference genome (NCBI Gene ID: 5542) encodes the medium allele. Regarding the primary structure of bPRP alleles, in the UniProtKB database (accession number P04280), the full amino-acid sequence of the large variant, deduced through experimental evidence at the protein level, is deposited.
2.1.4. Statherin and P-B Peptides
Statherin is encoded by the STATH gene localized on chromosome 4q13.3 [36]. It is a 43-amino-acid-residues multifunctional phospho-peptide characterized by an anomalous high content of tyrosine, proline and glutamine. The roles of statherin in the oral cavity are various. Due to its great affinity for calcium phosfate minerals, such as hydroxyapatite, it maintains the supersaturated state of calcium in human saliva, thus contributing to the mineral dynamics of the tooth surface and stabilizing the acquired enamel [37]. Furthermore, statherin is involved in bacterial colonization [38] and acts as a boundary lubricant on the enamel surface [39]. P-B is the product of the specific PROL3 gene located on chromosome 4q13.3, very close to the STATH gene [40]. Differently from bPRPs, which do not have aromatic amino acids in their sequences, and similarly to statherin, it has three tyrosine residues in its sequence. Furthermore, like statherin, the P-B peptide is secreted mainly by submandibular/sublingual (SM/SL) glands and also by parotids [41]. It is important to recall that P-B is also called, at the Swiss-Prot site (code P02814), “submaxillary gland androgen-regulated protein 3B precursor”, but researchers were not able to find any reference justifying the origin of this name. The specific role of the P-B peptide in saliva is still far from being clarified [42,43]. Nonetheless, its sequence resembles the ionic complement of statherin. Indeed, statherin has a small N-terminal negative tail followed by a long neutral domain with many proline residues, while the P-B peptide has a small N-terminal positive tail followed by a long neutral poly-proline domain. The two peptides could interact for the formation of the acquired enamel pellicle of the tooth. Different isoforms of statherin have been detected in human saliva; the variant called SV1 (Statherin variant 1) corresponds to statherin missing the C-terminal Phe residue (Statherin desPhe43) [41,44] (Figure 4a). The variant SV2, showing the deletion of the 6–15 internal residues with respect to statherin (Statherin des6-15), originates from alternative splicing that excludes an exon of 30 nucleotides [45]. SV3 corresponds to the SV2 variant lacking the C-terminal Phe residue. SV-1 lacking the C terminal Phe43 and statherin desThr42Phe43 (Figure 4a) was detected in parotid and submandibular secretory granules, suggesting that C-terminal removal occurs during the maturation of the secretory granules [6]. On the contrary, the N-terminal removal that generates statherin desAsp1 (Figure 4a) is probably an event occurring after granule secretion because this derivative was not detected in granules [6]. Statherin desAsp1 was also detected in human saliva by Vitorino and colleagues [46].

Figure 4. Pattern of proteoforms, mainly proteolytic fragments, originating from statherin (a), and the P-B peptide (b), characterized in human saliva by a top-down approach based on reversed-phase high pressure liquid chromatography-electrospray ionization tandem mass (RP-HPLC-ESI-MS/MS) analysis. The arrows indicate the main cleavage sites. The sequences recognized by furin-like convertase are evidenced in bold red, and the fragments derived by these cleavages are in shown in red.
Similarly, a small amount of P-B des1–5 was found in whole saliva as well as in parotid and submandibular-sublingual saliva but not in secretory granules, suggesting that this N-terminal cleavage also occurs after secretion [6]. Some P-B and statherin naturally occurring fragments are generated from a cleavage operated by a furin-like convertase and have been characterized by a top-down approach in researchers' previous studies [6,41]. Statherin undergoes convertase cleavages at the consensus sequence …R9R10IGR13…, generating statherin des1-9, des1-10, and des1-13 (Figure 4a), while P-B des1-5 is a convertase fragment of P-B peptide generated at the consensus sequence …R2GPR5… (Figure 4b). The P-B peptide undergoes further cleavages at the level of various chymotryptic-like sequences, generating several fragments detectable by a top-down proteomic approach [20] (Figure 4b).
2.1.5. Histatins (Hst)
Histatins are a family of small peptides deriving their name from the high number of histidine residues in their sequence. It is widely accepted that all the members of this family arise from two parent peptides, named histatin 1 and histatin 3, with very similar sequences and encoded by two genes (HIS1 and HIS2) located on chromosome 4q13 [47]. Despite the very high sequence similarity, these two peptides follow completely different PTM pathways. Histatin 3 (Hst3), differently from histatin 1 (Hst1), is submitted to a sequential cleavage, generating Hst6 (Hst3 1/25) at first, then Hst5 (Hst 3 1/24) subsequently, with powerful anti-fungal activity [48], and then other fragments [49]. Their different susceptibility to cleavage derives from the presence in Hst3 of the …RGYR↓… convertase consensus sequence, which is absent in Hst1 and thus is not cleaved. Hst1 today is considered a proangiogenic factor that may contribute to oral wound healing [50,51].
Some years ago, the group of Oppenheim [48] was able to establish the sequence of 12 histatins and named them from Hst1 to Hst12. The advent of the high-throughput MS apparatus applied to proteomic studies allowed for the determination of many other Hst fragments, the majority deriving from Hst3 [49,52]. For this reason, the nomenclature of histatins fragments has been recently modified and is reported on the Swiss Prot site with the code numbers P15515 (Hst1) and P15516 (Hst3).
2.1.6. Cystatins
Cystatins include type-1 cystatins (cystatins A and B), type-2 cystatins (C, D, S, SN, SA), and kininogens, or type-3 cystatins. Various biological activities have been demonstrated for these proteins, but a major role is linked to the inhibitory action exerted against cysteine proteinases. Thus, for this ability to modulate the proteolytic system, they are considered central in various diseases, including cancer [53,54,55,56]. Cystatins S, SN, SA, C, and D are encoded by loci CST1–5 closely clustered on chromosome 20p11.21. It has been observed that S-type cystatins (S, SN, and SA), present at higher concentrations in SM/SL secretion than in parotid saliva [6], were absent or present at very low concentrations from both parotid and SM/SL secretory granules. This finding suggested that the secretion of S-type cystatins is not granule-mediated [6]. It is relevant to remark that cystatin A and B are leaderless [57]. Cystatin C was found sporadically in parotids, SM/SL, and whole saliva, but it was absent from secretory granules, whereas cystatin D was not found in any sample. Thus, it was suggested that cystatins C and D may have a different origin than the other salivary cystatins. The presence in human saliva of truncated proteoforms of cystatins has been suggested by Lupi and colleagues, who observed by HPLC-ESI-MS several masses possibly related to N-terminally truncated cystatins in the acidic soluble fraction of human saliva [58]. Indeed, a subsequent in-depth study performed both in the acidic supernatant of whole saliva and in RP-HPLC-enriched fractions, by an integrated top-down/bottom-up pipeline, characterized some truncated proteoforms of cystatins [57]. The study evidenced that not all cystatins undergo proteolytic modifications. The following truncated proteoforms for the widespread cystatin SN and its natural variant SN Pro11 → Leu were detected and characterized: cystatin SN des1–4, and SN des1−7, cystatin SN Pro11 → Leu des1–4, and SN Pro11 → Leu des1–7. A truncated form of cystatin SA lacking the first seven amino acids from the N-terminus (cystatin SA des1–7) was also detected. Three truncated forms of the variant cystatin D Cys26 → Arg have been detected in human saliva, but none for the variant Arg26 → Cys. For easier reading, researchers will utilize the name cystatin D to refer to the variant Cys26 → Arg. Cystatin D des1–4 and des1–8 have been characterized by a top-down Fourier-transform ion cyclotron resonance mass-spectrometry pipeline [59] and also detected in the study of Manconi and colleagues [57] A third truncated proteoform of cystatin D lacking the first five amino-acid residues with the N-terminal glutamine converted to pyroglutamic acid (pGlu-cystatin D des1–5) was also characterized in the latter study. The authors speculated that the high abundance of the latter truncated proteoform was due to the greater resistance from degradation by amino peptidases due to N-terminal pyroglutamination. Cystatin B in adult human saliva is commonly detectable as an intact proteoform, and the two fragments 1-53 and 54-98 were characterized in the saliva of human preterm newborns [60].
2.2. Post-Secretory Cleavages
Almost all the families of salivary proteins and peptides described above (and others) are substrates of several proteolytic enzymes present in the mouth deriving from oral flora [61,62]. Since they are myriad, reporting a complete list is impossible, and in the following, researchers will refer to excellent published lists. Concerning PRPs and particularly bPRPs, many recent mass-spectrometric studies allowed us to report many bPRPs fragments [23,43,46,52], with high affinity for the tooth enamel [52]. However, the observation of a recurrent XPQ C-terminal sequence in many fragments detected induced the group of Oppenheim and Helmerhorst to characterize a glutamine endopeptidase from Rhotia sp [63]. Using the synthetic substrates KPQ-pNA and GGQ-pNA, the overall K(m) values were determined to be 97 +/− 7.7 and 611 +/− 28 micromolar, respectively, confirming glutamine endoprotease activity in whole saliva and the influence of the amino acids in positions P(2) and P(3) on protease recognition. The pH optimum of KPQ-pNA hydrolysis was 7.0, and the activity was most effectively inhibited by antipain and 4-(2-aminoethyl) benzene sulfonyl fluoride. The enzyme is metal-ion-dependent and not inhibited by cysteine protease inhibitors. A systematic evaluation of enzyme activities in various exocrine and non-exocrine contributors to whole saliva revealed that the glutamine endoprotease derives from Rhotia and is localized in dental plaque [63,64].
A further protein submitted to fragmentation in the mouth is the polymeric immunoglobulin receptor (PIgR), a type-I transmembrane glycoprotein playing the main role in the adaptive immune response on mucosal surfaces [65,66]. It transports polymeric IgA across mucosal epithelial cells. A proteolytic cleavage occurring in the glycosylated extra-cellular portion of pIgR generates the secretory component (19–603 residues), which has also been detected in human saliva [67]. The cleavage occurs by the action of unknown proteases, probably released by activated neutrophils [66], and the highly conserved sequence 602–613 (PRLFAEEKAVAD) is believed to be the cleavage signal [65]. Two peptides deriving from PIgR are detectable in saliva by a top-down proteomic approach; they are named AVAD and ASVD [68]. The peptide named AVAD originates by the cleavage occurring in this region at the level of Lys609, and the ASVD peptide derives from AVAD by the trypsin-like cleavage at Arg622. AVAD and ASVD peptides do not derive from the secretory component and have a sequence partially overlapped by the transmembrane portion (639–661) of PIgR. Thus, they should originate by degradation of PIgR after its release from disrupted cell membranes.
2.3. Proteolytic Cleavages: Variations Related to Age and Pathologies
Interestingly, it was observed that several proteolytic cleavages change according to age and in several diseases. Iavarone and colleagues [60] detected in the saliva of human preterm newborns sensible amounts of cystatin B fragments 1–53 and 54–98, suggesting in foetuses the presence of high-active specific proteolytic events that disappeared in adults. The cleavage involves a tyrosine and phenylalanine couple (Tyr↓Phe) and can be ascribed to a chymotrypsin-like enzymatic activity with strict specificity, as suggested by the very precise consensus sequence necessary for the cleavage. The two fragments were detected at very high relative amounts with respect to intact cystatin B in very preterm newborns, for which the concentration decreased as a function of the post-conceptional age (PCA) and they were practically undetectable when the age corresponded to that of full-term newborns. A search on the Merops protease database (release 12.4, accessed on 9 January 2017, http://merops.sanger.ac.uk/) returned various possibilities other than chymotrypsin A, such as chymosin, cathepsin E, metalloproteinase 2, ADAMTS4, endothelin converting enzyme 1, and some peptidases of the chymase class (mast cell chymotrypsin-like proteinase). It is impossible to establish if cystatin B is a natural substrate of the enzyme, thereby implying a functional role for the fragments as potential cryptides, or if the fragments observed are byproducts of a proteinase, without any functional meaning, their activity is devoted to other specific foetal oral cleavage processes. Similarly, an increased activity of convertases and carboxypeptidases responsible for the cleavages of aPRPs, histatins, and statherin was observed in preterm newborns with a low PCA of approximately 190 days. The activity decreased according to the PCA, and it reached the levels observed in the adult around the normal term of delivery [69]. Interestingly, this behavior was observed for the release of the PRP3-type aPRPs and the P-C peptide from PRP-1-type aPRPs, as well as for the release of histatin 6 and histatin 5 from histatin 3 and for the release of the fragments of statherin missing C-terminal residues. This was not the case for statherin desAsp1, which showed low levels or was not detectable in preterm newborns at low PCA but increased after birth, reaching values similar to those determined in at-term newborns [70]. Because statherin desAsp1 is a fragment of statherin not detectable in granule preparations [6], this result is a further clue that the proteinases (convertases and carboxypeptidases) involved in the cleavages of these families of salivary proteins are confined in the Golgi apparatus.
It has been reported that proteolytic fragmentations of salivary proteins may vary in some pathological states compared to physiological conditions. For instance, a study performed on a group of children and adolescents affected by type-1 diabetes revealed that several small peptides, most likely originated by post-secretory proteolytic cleavages occurring in the mouth, showed a higher concentration with respect to sex- and age-matched controls, suggesting increased activity of exogenous proteinases in the oral cavity of diabetics [70]. The peptides, characterized as fragments 1–14, 1–25, 5–25, 26–35, 26–44, and 36–44 of the P-C peptide, have been already detected in saliva from healthy subjects [63], and fragments 1–14 and 26–44 were also detected in the parotid saliva of healthy subjects by Hardt and colleagues [71]. Furthermore, an association of fragments 1–14 and 26–44 to high numbers of dental caries and the presence of fragments 1–14 in the saliva of subjects affected by Sjogren’s syndrome have been demonstrated by Huq and colleagues [72]. A top-down proteomic study investigated the salivary proteome of 49 multiple sclerosis (MuSc) patients and 54 healthy controls, quantifying 119 salivary peptides/proteins [73]. Among the observed differences, the fragments 1–14, 26–44, and 36–44 of the P-C peptide, the SV1 fragment of statherin, and cystatin SN des1–4 showed higher levels in patients with respect to controls. In a study performed to characterize possible differences in the salivary proteome of subjects affected by Wilson’s disease (WD) with respect to healthy controls, increased levels of the AVAD and ASVD fragments of PIgR (see above) were observed in patients, probably because of an increased disruption of cell membranes due to the high production of ROS typical of WD [68]. A higher level of the ASVD peptide was also determined in MuSc patients with respect to healthy controls [73]. Interestingly, a study performed on patients affected by systemic mastocytosis (SM) and grouped into SM with (SM+C) or without (SM-C) additional cutaneous lesions evidenced that the two SM forms were distinguished by the lower levels of PRP-3, PRP-3 desArg106, statherin desPhe43, P-B des1–5, and cystatin D des1–5 and des1–8 in SM-C patients with respect to SM+C [74]. It should be outlined that lower levels of cystatin D des1–5 and des1–8 have been also observed by comparing saliva of healthy elderly subjects with respect to adults [75].
The study was performed on patients affected by autoimmune hepatitis (AIH) and primary biliary cholangitis (PBC), two autoimmune liver diseases characterized by chronic hepatic inflammation and progressive liver fibrosis, to establish a panel of salivary proteins/peptides able to classify with good accuracy PBC patients vs HCs, AIH patients vs HCs, and PBC vs AIH patients. Among the other data, they revealed significantly different levels of PRP-3 desArg106 by comparing the two patient groups [76].
Higher levels of statherin desPhe43 and two others naturally occurring fragments (des1–9, and des1–13) were measured in the saliva of a group of Alzheimer’s disease patients compared to controls [77].
This entry is adapted from the peer-reviewed paper 10.3390/ijms241612776
This entry is offline, you can click here to edit this entry!
