Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Biology
Cytoplasmic phosphoinositides (PI) are critical regulators of the membrane–cytosol interface that control a myriad of cellular functions despite their low abundance among phospholipids. The metabolic cycle that generates different PI species is crucial to their regulatory role, controlling membrane dynamics, vesicular trafficking, signal transduction, and other key cellular events. The synthesis of phosphatidylinositol (3,4,5)-triphosphate (PI3,4,5P3) in the cytoplamic PI3K/Akt pathway is central to the life and death of a cell.
- phosphoinositide
- scaffolding protein
- PI3K-Akt pathway
1. Introduction
Autophagy is the “self-eating” of cytosolic components and is an essential cellular homeostasis mechanism under conditions of nutrient scarcity. It is a multi-step process, forming specific autophagic organelles in each step [125]. Organelles initiate from the membrane formation of omegasomes, gradually elongate to phagophores and enclose to generate an autophagosome, and then fuse with lysosome to form autolysosomes, which degrade the cargo. Phosphoinositide metabolism plays a key role in every step of autophagy. Different phosphoinositides recruit specific compartments on membrane structures, mediating the proper generation and activation of autophagic organelles [125,126] (Figure 1).
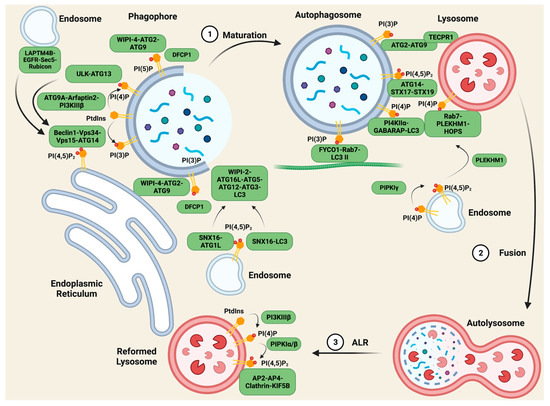
Figure 1. PIs regulate scaffolding complexes in autophagy. During the maturation of autophagosomes, the fusion between autophagosomes and lysosomes, and the reformation of lysosomes after degradation, membrane PIs play vital roles in recruiting specific scaffolding complexes to mediate lipid transfer, vesicle transport, and membrane tethering. The most important PI species in autophagy are PI(3)P and PI(4)P, as they anchor multiple crucial complexes in different steps of autophagy, while PI(5)P and PI(4,5)P2 also have unique roles. Figure created with biorender.com.
2. PI Complexes Regulate Autophagosome Maturation
The initiation of autophagic membrane structures is carried out by the protein Beclin1 [127,128,129]. Autophagy is stimulated by multiple stressors including serum starvation [130]. Upon serum starvation, inactive EGFR internalizes into LAPTM4B-positive endosomes [95]. Inactive EGFR and LAPTM4B stabilize each other at the late endosome and interact with multiple subunits of the exocyst complex, including Sec5. The EGFR-LAPTM4B-Sec5 complex recruits the autophagy inhibitor Rubicon, which in turn disassociates Rubicon from the Beclin1 complex and initiates the autophagy process [95]. The inactive EGFR-LAPTM4B complex specifically regulates autophagy, whereas the active EGF-EGFR complex prolongs EGFR signaling by blocking internalization into the lysosome where it is downregulated via the degradation of the receptor.
Beclin1 initiates the formation of the Beclin1-Vps34-Vps15 scaffolding complex, and the complex is recruited to the ER by ATG14 for autophagy specificity [131,132], while Vps34 generates PI(3)P from PtdIns. Vps34, and together with Vps15, Vps30, ATG14, and ATG38, forms PI3K complex I (PI3KCI) [131,133,134,135]. In yeast, PI3KCI constitutively binds to the vacuolar membrane protein Vac8 through ATG14 [136]. Upon autophagy initiation, ATG 1 recruits ATG9, ATG13, and ATG17-ATG31-ATG29 to assemble the ATG1 complex [137,138,139]. In an ATG1 kinase activity-dependent manner, PI3KCI associates with the ATG1 complex via the ATG38-ATG1 complex and Vps30-ATG9 interactions, anchoring PI3KC1 to the pre-autophagosomal structure for PI(3)P production [136]. A minor pool of PI(3)P is also generated from PI3KC2 in starvation-induced autophagy [22]. DFCP1, a protein containing the PI(3)P-binding FYVE domain, is recruited to these pools of PI(3)P, and together regulate the biogenesis of lipid droplets [140], acting as markers of the omegasomes [141].
PI(3)P also plays a critical role in the maturation of phagophores to autophagosomes. WIPI-2 binds to PI(3)P on the omegasomes and mediates ATG16L-ATG5-ATG12 complex formation [142]. The ATG16L complex acts together with ATG3 as an E3-like ligase and E2-enzyme for LC3 lipidation, coupling LC3 with phosphatidylethanolamine (PE) to generate LC3 II, which regulates the closure, fusion, and transport of the autophagosome [143,144]. Another PI(3)P effector, WIPI-4, forms a complex with ATG2 and ATG9 on the PI(3)P pools, tethering the autophagic membrane to the ER for lipid transfer to elongate this membrane structure [145].
Phosphoinositides are also involved in the initiation of autophagosomes. ATG9A, the ubiquitously expressed variant of ATG9, locates to autophagosomes on vesicle compartments where it binds to Arfaptin2 and PI4KIIIβ, bringing the complex to autophagic sites, and promoting the generation of PI(4)P [146]. PI(4)P then recruits the ULK-ATG13 complex, which activates Vps34 [146,147], enhancing autophagy as positive feedback. PI(5)P synthesized by PIKfyve on phagophores can act as a binding partner of DFCP1 and WIPI2 independent of the PI(3)P pool, forming scaffolding complexes and mediating the biogenesis of autophagy as mentioned above [148]. PI(4,5)P2 plays a more controversial role in autophagy initiation. On the one hand, PI(4,5)P2 phosphorylated from PI(5)P by PIPKIIs negatively regulates autophagosome biogenesis by depleting the PI(5)P pool. On the other hand, PI(4,5)P2 contributes to the autophagic membrane generated from endosomes and the ER. On ATG16L1-positive endosomes, SNX16, a PX-domain-containing protein, that specifically interacts with PI(4,5)P2, ATG16L1, and LC3, mediates the delivery of the ATG16L1 complex to the autophagic site and the lipidation of LC3 [149], while another pool of PI(4,5)P2 generated by PIPKIγi5 on the ER binds to ATG14, stabilizing the autophagy-specific Beclin-Vps34-Vps15-ATG14 scaffolding complex [150].
3. PI Complexes Regulate Autolysosome Fusion
After the maturation of the autophagosome, the next step of cellular content degradation is fusion with lysosomes, where phosphoinositide scaffolding complexes also play a vital role. To fuse with lysosomes, the autophagosome must first be transported by microtubules, and the tethering of the autophagosome and microtubule depends on FYCO1-Rab7-PI(3)P-LC3 II complex formation [151,152]. This allows the autophagosome to be transported to the cell periphery, where the lysosomes reside. Upon coming into proximity with the lysosomes, lysosome membrane protein TECPR1 interacts with autophagosome membrane proteins ATG5–ATG12, enabling its binding to autophagosome PI(3)P through its PH domain. This intervesicular complex facilitates lysosome–autophagosome fusion [153].
PI(4)P presented on both the autophagosome and lysosome membranes is crucial for their fusion. Gamma-aminobutyric acid receptor-associated protein (GABARAP) binds and recruits PI4KIIα to the autophagosome for PI(4)P generation. Knocking down this complex results in the enlargement of the autophagosome and the accumulation of IC3 II, indications of impaired autophagosome–lysosome fusion [154]. On the lysosomes, PI(4)P synthesis is controlled by PI4KIIIβ, and this PI(4)P pool interacts with Rab7, anchoring the PH domain-containing protein family member 1 (PLEKHM1) and the homotypic fusion and protein-sorting (HOPS) complex, and PLEKHM1 binds to the LC3/GABARAP complex via the LC3 interaction region [155]. In this way, the PI(4)P pool on both vesicles regulate the tethering and fusion of autophagosomes and lysosomes. Other studies indicate that PI(4,5)P2 also controls this process via multiple mechanisms. The conversion of PI(4)P into PI(4,5)P2 by PIPKIγ inactivates Rab7 and releases PLEKHM1 from endosomes, allowing the complex’s formation at autolysosome fusion sites [156], while ATG14, a PI(4,5)P2 effector, binds to the soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) domain of STX17 and SNAP29, stabilizing the STX17-SNAP29 binary t-SNARE complex on autophagosomes to facilitate the fusion [157].
4. PI Complexes Regulate Autophagic Lysosome Reformation
After degradation, autolysosomes are required to be recycled back into new lysosomes. If nutrients are scarce, this is achieved via a process called autophagic lysosome reformation (ALR). Studies show that a specific pool of PI(4,5)P2 mediates this process. PtdIns on the autolysosome is first phosphorylated by PI4KIIIβ to PI(4)P, to be used as a substrate for PIPKIα/β to generate PI(4,5)P2 [158,159]. The PI(4,5)P2 on the autolysosome surface recruits the adaptor proteins AP2 and AP4, and clathrin to induce membrane budding and tubule formation [159]. Then, the motor protein KIF5B interacts with PI(4,5)P2 in a clathrin-dependent manner, driving autolysosome tubulation and ALR [160].
This entry is adapted from the peer-reviewed paper 10.3390/biom13091297
This entry is offline, you can click here to edit this entry!
