1. Introduction
OMVs were discovered over 50 years ago, and research on them has progressed significantly, particularly in modifying their functionality to expand their application potential. This review focuses on using OMVs as biocatalysts—particularly their functionalization with enzymes and their applications. Functionalizing OMVs for biocatalytic targets involves modified enzymes, with the immobilization of enzymes being the most common and relevant approach. Immobilized enzymes are stronger and more resistant to environmental changes than free enzymes in solution [
20,
21]. Additionally, OMVs can transport enzymes to distant targets in a protected state [
22]. Therefore, the immobilization of enzymes enables the expansion of OMVs’ utility, while enhancing enzymes’ activity and stability.
There are several methods of enzyme immobilization, including depositing enzymes on solid matrices, adsorption on porous materials, immobilization via covalent bonds, affinity immobilization, and entrapment or encapsulation [
20,
23]. Each of these methods has its own advantages and disadvantages. Enzyme deposition on solid matrices and entrapment are the most commonly applied immobilization methods among these platforms. It is important to note that the chosen immobilization method significantly impacts the modified enzyme’s properties. For example, if the goal is to produce an enzyme with a thermally or mechanically stable surface, entrapment using OMVs is the appropriate immobilization method. On the other hand, enzyme deposition on solid matrices, such as enzyme display on the surface of OMVs, may be recommended if the substrate cannot penetrate the trapping matrix due to its limited properties. In relation to the above statements, the method of enzyme immobilization with the OMV platform is also closely related to the prospective applications of OMVs. In this review, we discuss recent research involving techniques pertaining to enzyme entrapment in OMVs and enzyme display on the surface of OMVs to elaborate on the method of enzyme immobilization with the OMV platform (
Table 1).
Table 1. Methods for immobilizing enzymes using the OMV platform.
| Enzyme Immobilization |
Superiority |
Deficiency |
Detail Method/Illustration |
Application |
Ref. |
|
|
| Fusion with ClyA (and other OMVs’ anchoring motifs, such as OmpA) |
Numerous outer membrane proteins can serve as anchoring motifs |
Limited expression of proteins that are too large to be transported out of the cytoplasm |
 |
Degradation of paraoxon and antibiotics |
[7] |
| Bioconjugation with SpyTag/SpyCatcher |
Capable of displaying functional proteins that are challenging to export from the cell |
SpyTag’s and SpyCatcher’s covalent bond is irreversible. |
 |
Little is known regarding the use of Spy systems to display enzymes on OMVs for biocatalyst purposes |
* |
| Utilization of ice-nucleation protein (INP) |
Facilitating assemblies with trivalent scaffolds is highly promising for simultaneously expressing multiple enzymes for cascade reactions |
The size of INP is relatively bigger than Spycatcher. |
 |
Enhanced glucose yield from cellulose degradation |
[24,25] |
 |
Paraoxon degradation to para-nitrophenol |
[25] |
|
|
| Physical functionalization |
Comparatively simple operation |
Limited loading efficiency |
- -
-
Incubation
- -
-
Electroporation
- -
-
Sonication
- -
-
Extrusion
|
There has been no report on the use of this technique for enzyme entrapment in the OMVs lumen for biocatalytic purposes |
|
| Genetic engineering |
Improved enzyme stability and offers high loading efficiency |
Limited interaction with the surrounding substrate |
- -
-
The use of SpyTag/SpyCatcher 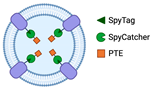
|
Decontamination of CWA, including paraoxon |
[16,17,18] |
- -
-
Direct binding to Lpp-OmpA 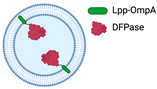
|
Hydrolyze DFP and paraoxon |
[19] |
- -
-
Fusion with Tat secretion signal peptide
|
GFP entrapment in OMVs lumen (there have been no data for enzyme entrapment utilizes Tat signal for functionalization of OMVs as biocatalysts) |
[26] |
- -
-
Fusion with Sec secretion signal peptide
|
Bioconversion of fatty acid |
[27] |
2. Enzyme Display on OMV Surface
2.1. Fusion with ClyA
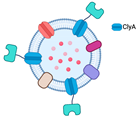
The recent prevalence of OMV surface decoration may be traced back to one of the earliest investigations of Kim et al. [
7]. That study functionalized the OMV surface using fusion binding with Cytolysin A (ClyA), a pore-forming hemolytic protein (
Table 1). ClyA, initially identified in the
Escherichia coli K-12 strain, is known to be one of the periplasmic proteins secreted along with the vesicles from the bacterial cell, suggesting its use in a vesicle transport mechanism in bacteria [
6,
28,
29,
30]. The expression of the fusion protein with ClyA outside the cell indicates that this porin can serve as an anchoring motif. While this study was one of the pioneer investigations into the use of OMVs as vaccines, the concept’s essence can also be applied to displaying enzymes, thereby broadening the use of OMVs as biocatalysts.
Kim et al. conducted experiments in which they fused several heterologous proteins to the ClyA toxin with N-terminus and C-terminus variations. The proteins β-lactamase (Bla), organophosphorus hydrolase (OPH), and β-galactosidase (LacZ) were coupled with ClyA to produce synthetic OMVs. Before combining ClyA with the enzymes, they fused it with GFP to assess the effect of fusion on the N- and C-termini. The combination of ClyA with GFP, Bla, and OPH, where ClyA was bound to the N-terminus fusion protein, showed a considerable increase in cell-surface and vesicle activity. On the other hand, Bla bound to the N-terminus of ClyA resulted in a significant decrease in activity. Similarly, ClyA bound to the C-terminus of OPH had no measurable effect on OPH activity. Given that the OPH substrate used in the study, paraoxon, is membrane impermeable, OPH bound outside the cell or vesicle was preferred [
7]. In line with this hypothesis, Kim et al. also fused LacZ to the C-terminus of the ClyA anchor. Their research suggested that ClyA could serve as an anchoring motif for decorating the surface of OMVs with other proteins. In addition to ClyA, more than 60 membrane proteins have been identified as OMV anchoring proteins, including OmpA [
9,
31,
32,
33]. Further investigation of OMVs may uncover additional proteins with essential functions.
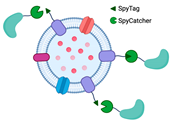
2.2. Bioconjugation with SpyTag/SpyCatcher
The SpyTag/SpyCatcher system has emerged as a powerful tool for the covalent bioconjugation of proteins. SpyTag and SpyCatcher are derived from the CnaB2 domain of the fibronectin-binding protein FbaB of
Streptococcus pyogenes. SpyTag is a small peptide consisting of 13 amino acids, while SpyCatcher is a larger protein fragment containing 116 residues. This system is based on the covalent peptide interactions between SpyTag and SpyCatcher protein pairs, which result in the formation of an irreversible isopeptide bond between lysine and aspartic acid residues. The isopeptide bond formation is spontaneous and occurs within minutes, without the need for specific pH or reaction temperature conditions. In addition, it is compatible with various buffers [
34,
35,
36]. The simplicity and versatility of this system have made it a popular choice for protein ligation studies, including antibody targeting in vaccine development [
37,
38] and reporter conjugation in biosensor [
39] applications.
SpyTag and SpyCatcher can be fused at either the N-terminus or the C-terminus. In addition to directly fusing the functional protein to the anchoring motif, the use of SpyTag or SpyCatcher to fuse with the C-terminus of the outer membrane protein is a way of functionalizing OMVs through surface display. One advantage of using SpyTag or SpyCatcher to fuse with the C-terminus of an outer membrane protein is the ability to display proteins that are challenging to transport out of the cell, such as large proteins or those with limited expression levels in the cytoplasm. To conjugate the fused protein with SpyTag or SpyCatcher, the only requirement is adding the fused protein to the buffer. For instance, the SpyTag/SpyCatcher system was used to display the virulence factor hemoglobin protease (Hbp) on the OMV platform to investigate optimal exposure of multiple antigenic sites to the immune system [
37]. The Hbp–SpyTag display was achieved by fusing Hbp to the C-terminus of SpyTag, which enabled the stable attachment of various antigens to Hbp. In this case, SpyTag expressed far from the surface of OMVs provide broad access without steric hindrance for conjugation with its fusion partner, SpyCatcher, suggesting the potential for more stable attachment of various antigens to Hbp. The SpyTag/SpyCatcher system has also been employed to display the receptor binding domain (RBD) of the SARS-CoV-2 spike protein on the surface of OMVs. That study demonstrated the potential of the SpyTag/SpyCatcher system for displaying complex proteins and suggested its potential use for vaccine development against COVID-19 [
38]. Although using the Spy toolbox for functionalizing OMVs as a biocatalyst is uncommon, the concept should inspire future development of OMVs for biocatalytic requirements. The SpyTag/SpyCatcher system offers a simple and efficient method for displaying proteins on the surface of OMVs (
Table 1), providing a promising avenue for developing new biocatalysts.
Sometimes the functionalization of OMVs for biocatalyst purposes using the Spy system does not require engineering of the OMV surface. Instead, SpyTag is conjugated to the transmembrane porin protein OmpA at the internal, N-terminus, and C-terminus regions to create an entrapment platform for OMVs. For example, the phosphotriesterase (PTE) enzyme is fused with SpyCatcher, and the sequence leader torA is added to localize PTE-SpyCatcher expression in the periplasm. This allows the enzymes to be encapsulated within the OMVs [
16,
17]. This research demonstrates that applying the Spy tools is highly adaptable to achieve desired architectural structures and functions, and that it is not restricted to modifying OMV surfaces.
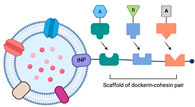
2.3. The Utilization of the Ice-Nucleation Protein
The ice-nucleation protein (INP) is found in bacteria that actively nucleate ice, such as
Pseudomonas syringae [
40],
Pseudomonas fluorescens [
41], and
Erwinia herbicola [
42]. It can speed up the formation of ice crystals in supercooled water [
43,
44]. This protein comprises over 1200 amino acid residues and consists of three domains: the N-terminus domain, the central cylindrical repeating domain (CRD), and the C-terminus domain. The N-terminus domain is hydrophobic, while the C-terminus domain is hydrophilic. The N-terminus acts as a membrane anchor for the extracellularly expressed C-terminus; however, proteins fused to either or both termini are exposed on the cell surface. The CRD is a catalytic domain that contributes to the formation of ice crystals and is unrelated to anchoring motifs. However, it functions as a distance regulator between heterologous proteins and the cell surface [
45,
46].
Enzymes can be effectively immobilized on the outer membrane surface using the INP (
Table 1). The INP has the ability to display the trivalent scaffold, Scaf3, on the bacteria cell surface by generating the INP-scaf3 fusion by engineering the episomal DNA [
24,
47]. Park et al. used the INP-scaf3 plasmid, containing three different cohesin domains, to immobilize multiple enzymes on the surface of OMVs simultaneously in the hyper-vesiculating mutant
E. coli JC8031. This configuration allows the dockerin of each of the three different cellulases to interact with their respective cohesin partners, thereby providing a promising approach for the simultaneous and site-specific expression of multiple enzymes that are useful for cascade reactions [
24].
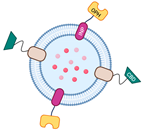
Similarly, Su et al. reported using a different design to engineer the surface of OMVs using the INP. In their study, they fused the OPH enzyme directly to the INP in the pVLT33-INPOPH6 construct to immobilize the enzyme on the surface of OMVs. Additionally, they fused cellulose binding domains (CBD) with the
E. coli lipidation sequence of major lipoprotein-outer membrane protein A (Lpp-OmpA) to form the pUCBD plasmid, in which Lpp-OmpA was used as an anchoring motif. This approach allows the two engineered proteins to be expressed outside of the cell, with the CBD providing an advantage in terms of OMV recovery and enabling the synthetic OMV-enzymes to be utilized multiple times [
25].
Both studies demonstrated that the INP offers construction flexibility for displaying diverse functional proteins on OMV platforms and facilitates recovery and purification. Thus, this anchoring motif represents a promising tool for future biotechnological applications in enzyme immobilization.
3. OMV-Mediated Encapsulation
Encapsulation does not provide flexible interaction and high mass transfer between proteins and their environment. However, the encapsulation technique offers enzyme protection and increases enzyme stability by shielding enzymes from harsh environments, since the matrices will carry them in the lumen to travel long distances. The use of OMVs as the encapsulation matrices is more commonly applied in biomedicine for loading small molecules, such as pharmaceutical compounds [
48,
49]. However, this does not preclude its application for the biocatalytic functionalization of OMVs. Huang et al. categorized the functionalization of OMVs into (i) physical or chemical approaches and (ii) genetic engineering [
50]. However, in this article, we do not discuss the functionalization of OMVs through a chemical approach—for instance, the 1-ethyl-3-[3-dimethylaminopropyl]-carbodiimide hydrochloride/
N-hydroxysuccinimide (EDC/NHS) coupling reaction—as it is not a material encapsulation technique used for functionalized OMVs as biocatalysts. In addition, we examine the genetic-engineering-based functionalization of OMVs specifically for the encapsulation technique.
3.1. Physical Funct3ionalization
Incubation is the simplest way to introduce biomolecules or functional proteins into the lumen of OMVs. However, the loading efficiency is often uncontrolled, requiring large quantities of loading material to be incubated with the growth media. Alternatively, physical functionalization methods, such as electroporation, sonication, and extrusion, have been explored but require greater effort [
50,
51,
52,
53] For instance, electroporation has encapsulated gold nanoparticles (AuNPs) [
51]. However, the use of this method for enzyme encapsulation and for utilizing OMVs as biocatalysts has yet to be documented, probably because it is difficult to prepare enzymes in large quantities. Another challenge is that the cargo must be delivered to the lumen while maintaining its enzymatic activity.
3.2. Genetic Engineering Approach
Most OMVs used as biocatalysts are functionalized using genetic engineering approaches, such as the Spy toolbox mentioned earlier (
Table 1). The idea of incorporating heterologous proteins into the lumen of OMVs was first explored by Kesty et al., who expressed GFP fused to the Tat signal peptide. This led to the detection of GFP in both the periplasm and OMVs, suggesting that OMVs partially enclose periplasmic contents and transport them outside the cell [
26]. Later, the Spy system was developed, which utilized the SpyTag/SpyCatcher pair to bind the PTE enzyme to OmpA on the inner side of the OMV membrane [
16,
17,
18]. In addition to bioconjugation with the Spy pair bound to the outer membrane protein OmpA, an alternative encapsulation method involves direct binding to the N-terminus of the lipidation sequence (Lpp) (
Table 1), which has been demonstrated to hold the diisopropyl fluorophosphatase (DFPase) enzyme under the lipid bilayer [
19].
Recent studies have shown the potential of signal peptides for localizing fatty acid-converting enzyme expression in the periplasm. While the Tat signal was previously used for this purpose, the enzyme is now secreted into the periplasm via the Sec pathway, guided by the PelB secretion signal peptide (PelBSS) from pectate lyase B of
Erwinia carotovora. Fatty acid double bond hydratase from
Stenotrophomonas maltophilia (SmOhyA) and photoactivated fatty acid decarboxylase from
Chlorella variabilis NC64 A (CvFAP) were fused with PelBSS to allow for periplasmic expression, which was subsequently encased by OMVs [
27]. Similarly, in another study, the β-lactamase enzyme encoded by the CMY-10 gene was secreted into the periplasm using PelBSS. Both studies employed a hyper-vesiculating
E. coli mutant lacking the
tolA and
tolR genes [
21]. The outer membrane protein serves as an anchoring motif for displaying enzymes or functional proteins extracellularly and as a holding motif for the loading material beneath the lipid bilayer during encapsulation. Moreover, since OMVs contain periplasmic contents, localizing loading material with signal peptides that can penetrate the inner membrane and accumulate in the periplasm significantly aids in immobilizing enzymes by encapsulating them using the OMV platform.
Overall, using signal peptides for enzyme localization and encapsulation within OMVs represents a promising approach for developing novel biocatalysts and drug delivery systems. While more research is needed to optimize the efficiency and specificity of this approach, it has the potential to revolutionize the field of biotechnology and to open new avenues for developing advanced therapeutics.
This entry is adapted from the peer-reviewed paper 10.3390/membranes13050459






